Targeting colorectal cancer with small-molecule inhibitors of ALDH1B1
- PMID: 35788181
- PMCID: PMC9529790
- DOI: 10.1038/s41589-022-01048-w
Targeting colorectal cancer with small-molecule inhibitors of ALDH1B1
Erratum in
-
Author Correction: Targeting colorectal cancer with small-molecule inhibitors of ALDH1B1.Nat Chem Biol. 2025 Feb;21(2):309. doi: 10.1038/s41589-024-01810-2. Nat Chem Biol. 2025. PMID: 39653787 No abstract available.
Abstract
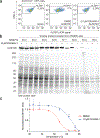
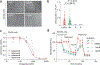
Aldehyde dehydrogenases (ALDHs) are promising cancer drug targets, as certain isoforms are required for the survival of stem-like tumor cells. We have discovered selective inhibitors of ALDH1B1, a mitochondrial enzyme that promotes colorectal and pancreatic cancer. We describe bicyclic imidazoliums and guanidines that target the ALDH1B1 active site with comparable molecular interactions and potencies. Both pharmacophores abrogate ALDH1B1 function in cells; however, the guanidines circumvent an off-target mitochondrial toxicity exhibited by the imidazoliums. Our lead isoform-selective guanidinyl antagonists of ALDHs exhibit proteome-wide target specificity, and they selectively block the growth of colon cancer spheroids and organoids. Finally, we have used genetic and chemical perturbations to elucidate the ALDH1B1-dependent transcriptome, which includes genes that regulate mitochondrial metabolism and ribosomal function. Our findings support an essential role for ALDH1B1 in colorectal cancer, provide molecular probes for studying ALDH1B1 functions and yield leads for developing ALDH1B1-targeting therapies.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
J.K.C., Z.F., M.E.H., T.K., C.R.M., and A.E.O. have filed a PCT application related to the compounds described in this study.
Figures











References
METHODS-ONLY REFERENCES
-
- Taipale J et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406, 1005–1009, (2000). - PubMed
-
- Hoppel C, DiMarco JP & Tandler B Riboflavin and rat hepatic cell structure and function. Mitochondrial oxidative metabolism in deficiency states. J. Biol. Chem 254, 4164–4170, (1979). - PubMed
-
- Elias JE & Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214, (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

