Y RNAs are conserved endogenous RIG-I ligands across RNA virus infection and are targeted by HIV-1
- PMID: 35789859
- PMCID: PMC9250025
- DOI: 10.1016/j.isci.2022.104599
Y RNAs are conserved endogenous RIG-I ligands across RNA virus infection and are targeted by HIV-1
Abstract
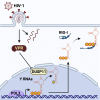
Pattern recognition receptors (PRRs) protect against microbial invasion by detecting specific molecular patterns found in pathogens and initiating an immune response. Although microbial-derived PRR ligands have been extensively characterized, the contribution and relevance of endogenous ligands to PRR activation remains overlooked. Here, we characterize the landscape of endogenous ligands that engage RIG-I-like receptors (RLRs) upon infection by different RNA viruses. In each infection, several RNAs transcribed by RNA polymerase III (Pol3) specifically engaged RLRs, particularly the family of Y RNAs. Sensing of Y RNAs was dependent on their mimicking of viral secondary structure and their 5'-triphosphate extremity. Further, we found that HIV-1 triggered a VPR-dependent downregulation of RNA triphosphatase DUSP11 in vitro and in vivo, inducing a transcriptome-wide change of cellular RNA 5'-triphosphorylation that licenses Y RNA immunogenicity. Overall, our work uncovers the contribution of endogenous RNAs to antiviral immunity and demonstrates the importance of this pathway in HIV-1 infection.
Keywords: Biological sciences; Immunology; Transcriptomics.
© 2022 The Authors.
Conflict of interest statement
The authors declare no conflicting interests.
Figures




References
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. - DOI
-
- Burke J.M., Kincaid R.P., Nottingham R.M., Lambowitz A.M., Sullivan C.S. DUSP11 activity on triphosphorylated transcripts promotes Argonaute association with noncanonical viral microRNAs and regulates steady-state levels of cellular noncoding RNAs. Genes Dev. 2016;30:2076–2092. doi: 10.1101/gad.282616.116. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

