Evaluating Automatic Segmentation for Swallowing-Related Organs for Head and Neck Cancer
- PMID: 35790457
- PMCID: PMC9340321
- DOI: 10.1177/15330338221105724
Evaluating Automatic Segmentation for Swallowing-Related Organs for Head and Neck Cancer
Abstract
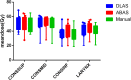
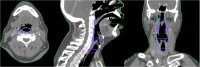
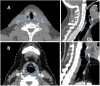
Purpose: To evaluate the accuracy of deep-learning-based auto-segmentation of the superior constrictor, middle constrictor, inferior constrictor, and larynx in comparison with a traditional multi-atlas-based method. Methods and Materials: One hundred and five computed tomography image datasets from 83 head and neck cancer patients were retrospectively collected and the superior constrictor, middle constrictor, inferior constrictor, and larynx were analyzed for deep-learning versus multi-atlas-based segmentation. Eighty-three computed tomography images (40 diagnostic computed tomography and 43 planning computed tomography) were used for training the convolutional neural network, and for atlas-based model training. The remaining 22 computed tomography datasets were used for validation of the atlas-based auto-segmentation versus deep-learning-based auto-segmentation contours, both of which were compared with the corresponding manual contours. Quantitative measures included Dice similarity coefficient, recall, precision, Hausdorff distance, 95th percentile of Hausdorff distance, and mean surface distance. Dosimetric differences between the auto-generated contours and manual contours were evaluated. Subjective evaluation was obtained from 3 clinical observers to blindly score the autosegmented structures based on the percentage of slices that require manual modification. Results: The deep-learning-based auto-segmentation versus atlas-based auto-segmentation results were compared for the superior constrictor, middle constrictor, inferior constrictor, and larynx. The mean Dice similarity coefficient values for the 4 structures were 0.67, 0.60, 0.65, and 0.84 for deep-learning-based auto-segmentation, whereas atlas-based auto-segmentation has Dice similarity coefficient results at 0.45, 0.36, 0.50, and 0.70, respectively. The mean 95th percentile of Hausdorff distance (cm) for the 4 structures were 0.41, 0.57, 0.59, and 0.54 for deep-learning-based auto-segmentation, but 0.78, 0.95, 0.96, and 1.23 for atlas-based auto-segmentation results, respectively. Similar mean dose differences were obtained from the 2 sets of autosegmented contours compared to manual contours. The dose-volume discrepancies and the average modification rates were higher with the atlas-based auto-segmentation contours. Conclusion: Swallowing-related structures are more accurately generated with DL-based versus atlas-based segmentation when compared with manual contours.
Keywords: atlas-based auto-contouring; deep learning convolutional neural network; head and neck cancer; radiotherapy contouring; swallow-related organs.
Conflict of interest statement
Figures




References
-
- Gregoire V, Langendijk JA, Nuyts S. Advances in radiotherapy for head and neck cancer. J Clin Oncol. 2015;33(29):3277-3284. - PubMed
-
- Geets X, Daisne JF, Arcangeli S, et al. Inter-observer variability in the delineation of pharyngo-laryngeal tumor, parotid glands and cervical spinal cord: comparison between CT-scan and MRI. Radiother Oncol. 2005;77(1):25-31. - PubMed
-
- Eisbruch A, Schwartz M, Rasch C, et al. Dysphagia and aspiration after chemoradiotherapy for head-and-neck cancer: which anatomic structures are affected and can they be spared by IMRT? Int J Radiat Oncol Biol Phys. 2004;60(5):1425-1439. - PubMed
-
- Levendag PC, Teguh DN, Voet P, et al. Dysphagia disorders in patients with cancer of the oropharynx are significantly affected by the radiation therapy dose to the superior and middle constrictor muscle: a dose-effect relationship. Radiother Oncol. 2007;85(1):64-73. - PubMed
-
- Vorwerk H, Zink K, Schiller R, et al. Protection of quality and innovation in radiation oncology: the prospective multicenter trial the German society of radiation oncology (DEGRO-QUIRO study). Evaluation of time, attendance of medical staff, and resources during radiotherapy with IMRT. Strahlenther Onkol. 2014;190(5):433-443. - PubMed

