CRISPR/Cas9-mediated LINC00511 knockout strategies, increased apoptosis of breast cancer cells via suppressing antiapoptotic genes
- PMID: 35790898
- PMCID: PMC9254607
- DOI: 10.1186/s12575-022-00171-1
CRISPR/Cas9-mediated LINC00511 knockout strategies, increased apoptosis of breast cancer cells via suppressing antiapoptotic genes
Abstract
Background: The growing detection of long noncoding RNAs (lncRNAs) required the application of functional approaches in order to provide absolutely precise, conducive, and reliable processed information along with effective consequences. We utilized genetic knockout (KO) techniques to ablate the Long Intergenic Noncoding RNA 00,511 gene in several humans who suffered from breast cancer cells and at the end we analyzed and examined the results.
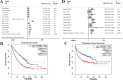
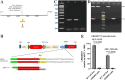
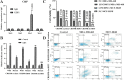
Results: The predictive relevance of LINC00511 expression pattern was measured by using a pooled hazard ratio (HR) with a 95% confidence interval (CI). The link among LINC00511 expression profiles and cancer metastasis was measured by using a pooled odds ratio (OR) with a 95% confidence interval. This meta- analysis was composed of fifteen studies which contained a total of 1040 tumor patients. We used three distinct CRISPR/Cas9-mediated knockdown techniques to prevent the LINC00511 lncRNA from being transcribed. RT-PCR was used to measure lncRNA and RNA expression. We used CCK-8, colony formation tests, and the invasion transwell test to measure cell proliferation and invasion. The stemness was measured by using a sphere-formation test. To validate molecular attachment, luciferase reporter assays were performed. The functional impacts of LINC00511 gene deletion in knockdown breast cancer cell lines were confirmed by using RT-qPCR, MTT, and a colony formation test. This meta-analysis was composed of 15 trials which contained a total of 1040 malignant tumors. Greater LINC00511 expression was ascribed to a lower overall survival (HR = 1.93, 95% CI 1.49-2.49, < P 0.001) and to an increased proportion of lymph node metastasis (OR = 3.07, 95% CI 2.23-4.23, P < 0.001) in the meta-analysis. It was found that the role of LINC00511 was overexpressed in breast cancer samples, and this overexpression was ascribed to a poor prognosis. The gain and loss-of-function tests demonstrated findings such as LINC00511 increased breast cancer cell proliferation, sphere-forming ability, and tumor growth. Additionally, the transcription factor E2F1 binds to the Nanog gene's promoter site to induce transcription. P57, P21, Prkca, MDM4, Map2k6, and FADD gene expression in the treatment group (LINC00511 deletion) was significantly higher than in the control group (P < 0.01). In addition, knockout cells had lower expression of BCL2 and surviving genes than control cells P < 0.001). In each of the two target alleles, the du-HITI approach introduced a reporter and a transcription termination signal. This strategy's donor vector preparation was significantly easier than "CRISPR HDR," and cell selection was likewise much easier than "CRISPR excision." Furthermore, when this approach was used in the initial transfection attempt, single-cell knockouts for both alleles were generated.
Conclusions: The methods employed and described in this work could be extended to the production of LINC00511 knockout cell lines and, in theory, to the deletion of other lncRNAs to study their function.
Keywords: Breast cancer; CRISPR/Cas9; Knockout; LINC00511.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

