Characterisation of Staphylococci species from neonatal blood cultures in low- and middle-income countries
- PMID: 35790903
- PMCID: PMC9254428
- DOI: 10.1186/s12879-022-07541-w
Characterisation of Staphylococci species from neonatal blood cultures in low- and middle-income countries
Abstract
Background: In low- and middle-income countries (LMIC) Staphylococcus aureus is regarded as one of the leading bacterial causes of neonatal sepsis, however there is limited knowledge on the species diversity and antimicrobial resistance caused by Gram-positive bacteria (GPB).
Methods: We characterised GPB isolates from neonatal blood cultures from LMICs in Africa (Ethiopia, Nigeria, Rwanda, and South Africa) and South-Asia (Bangladesh and Pakistan) between 2015-2017. We determined minimum inhibitory concentrations and performed whole genome sequencing (WGS) on Staphylococci isolates recovered and clinical data collected related to the onset of sepsis and the outcome of the neonate up to 60 days of age.
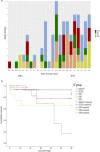
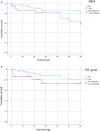
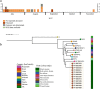
Results: From the isolates recovered from blood cultures, Staphylococci species were most frequently identified. Out of 100 S. aureus isolates sequenced, 18 different sequence types (ST) were found which unveiled two small epidemiological clusters caused by methicillin resistant S. aureus (MRSA) in Pakistan (ST8) and South Africa (ST5), both with high mortality (n = 6/17). One-third of S. aureus was MRSA, with methicillin resistance also detected in Staphylococcus epidermidis, Staphylococcus haemolyticus and Mammaliicoccus sciuri. Through additional WGS analysis we report a cluster of M. sciuri in Pakistan identified between July-November 2017.
Conclusions: In total we identified 14 different GPB bacterial species, however Staphylococci was dominant. These findings highlight the need of a prospective genomic epidemiology study to comprehensively assess the true burden of GPB neonatal sepsis focusing specifically on mechanisms of resistance and virulence across species and in relation to neonatal outcome.
Keywords: Early onset; Epidemiology; Genomics; LMIC; Late onset; Mammaliicocci; Mortality; Neonatal sepsis; Staphylococci.
© 2022. The Author(s).
Conflict of interest statement
The authors have no competing interests to declare.
Figures


References
-
- Okomo U, Akpalu ENK, le Doare K, Roca A, Cousens S, Jarde A, et al. Articles Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa : a systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect Dis. 2019;3099(19):1–16. doi: 10.1016/S1473-3099(19)30414-1. - DOI - PubMed
-
- Naghavi M, Abajobir AA, Abbafati C, Abbas KM, Abd-Allah F, Abera SF, et al. Global, regional, and national age-sex specifc mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100):1151–1210. doi: 10.1016/S0140-6736(17)32152-9. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

