Translation of cytoplasmic UBA1 contributes to VEXAS syndrome pathogenesis
- PMID: 35793467
- PMCID: PMC9523373
- DOI: 10.1182/blood.2022016985
Translation of cytoplasmic UBA1 contributes to VEXAS syndrome pathogenesis
Abstract
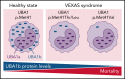
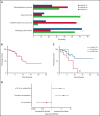
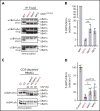
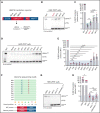
Somatic mutations in UBA1 cause vacuoles, E1 ubiquitin-activating enzyme, X-linked, autoinflammatory somatic (VEXAS) syndrome, an adult-onset inflammatory disease with an overlap of hematologic manifestations. VEXAS syndrome is characterized by a high mortality rate and significant clinical heterogeneity. We sought to determine independent predictors of survival in VEXAS and to understand the mechanistic basis for these factors. We analyzed 83 patients with somatic pathogenic variants in UBA1 at p.Met41 (p.Met41Leu/Thr/Val), the start codon for translation of the cytoplasmic isoform of UBA1 (UBA1b). Patients with the p.Met41Val genotype were most likely to have an undifferentiated inflammatory syndrome. Multivariate analysis showed ear chondritis was associated with increased survival, whereas transfusion dependence and the p.Met41Val variant were independently associated with decreased survival. Using in vitro models and patient-derived cells, we demonstrate that p.Met41Val variant supports less UBA1b translation than either p.Met41Leu or p.Met41Thr, providing a molecular rationale for decreased survival. In addition, we show that these 3 canonical VEXAS variants produce more UBA1b than any of the 6 other possible single-nucleotide variants within this codon. Finally, we report a patient, clinically diagnosed with VEXAS syndrome, with 2 novel mutations in UBA1 occurring in cis on the same allele. One mutation (c.121 A>T; p.Met41Leu) caused severely reduced translation of UBA1b in a reporter assay, but coexpression with the second mutation (c.119 G>C; p.Gly40Ala) rescued UBA1b levels to those of canonical mutations. We conclude that regulation of residual UBA1b translation is fundamental to the pathogenesis of VEXAS syndrome and contributes to disease prognosis.
© 2022 by The American Society of Hematology.
Figures
Comment in
-
Lost in translation: cytoplasmic UBA1 and VEXAS syndrome.Blood. 2022 Sep 29;140(13):1455-1457. doi: 10.1182/blood.2022017560. Blood. 2022. PMID: 36173661 No abstract available.
References
-
- Georgin-Lavialle S, Terrier B, Guedon AF, et al. ; French VEXAS group; GFEV, GFM, CEREMAIA, MINHEMON . Further characterization of clinical and laboratory features in VEXAS syndrome: large-scale analysis of a multicentre case series of 116 French patients. Br J Dermatol. 2022;186(3):564-574. - PubMed
-
- Bourbon E, Heiblig M, Gerfaud Valentin M, et al. Therapeutic options in VEXAS syndrome: insights from a retrospective series. Blood. 2021;137(26):3682-3684. - PubMed
-
- Tsuchida N, Kunishita Y, Uchiyama Y, et al. Pathogenic UBA1 variants associated with VEXAS syndrome in Japanese patients with relapsing polychondritis. Ann Rheum Dis. 2021;80(8):1057-1061. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

