Single-cell roadmap of human gonadal development
- PMID: 35794482
- PMCID: PMC9300467
- DOI: 10.1038/s41586-022-04918-4
Single-cell roadmap of human gonadal development
Abstract
Gonadal development is a complex process that involves sex determination followed by divergent maturation into either testes or ovaries1. Historically, limited tissue accessibility, a lack of reliable in vitro models and critical differences between humans and mice have hampered our knowledge of human gonadogenesis, despite its importance in gonadal conditions and infertility. Here, we generated a comprehensive map of first- and second-trimester human gonads using a combination of single-cell and spatial transcriptomics, chromatin accessibility assays and fluorescent microscopy. We extracted human-specific regulatory programmes that control the development of germline and somatic cell lineages by profiling equivalent developmental stages in mice. In both species, we define the somatic cell states present at the time of sex specification, including the bipotent early supporting population that, in males, upregulates the testis-determining factor SRY and sPAX8s, a gonadal lineage located at the gonadal-mesonephric interface. In females, we resolve the cellular and molecular events that give rise to the first and second waves of granulosa cells that compartmentalize the developing ovary to modulate germ cell differentiation. In males, we identify human SIGLEC15+ and TREM2+ fetal testicular macrophages, which signal to somatic cells outside and inside the developing testis cords, respectively. This study provides a comprehensive spatiotemporal map of human and mouse gonadal differentiation, which can guide in vitro gonadogenesis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


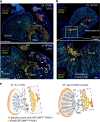











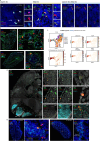
Comment in
-
Mapping gonadogenesis in time and space.Nat Rev Urol. 2022 Oct;19(10):577. doi: 10.1038/s41585-022-00652-8. Nat Rev Urol. 2022. PMID: 36028718 No abstract available.
References
-
- Hanley, N. A. et al. SRY, SOX9, and DAX1 expression patterns during human sex determination and gonadal development. Mech. Dev.91, 403–407 (2000). - PubMed
-
- Albrecht, K. H. & Eicher, E. M. Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev. Biol.240, 92–107 (2001). - PubMed
-
- Nef, S., Stévant, I. & Greenfield, A. Characterizing the bipotential mammalian gonad. Curr. Top. Dev. Biol.134, 167–194 (2019). - PubMed
-
- Maheshwari, A. & Fowler, P. A. Primordial follicular assembly in humans – revisited. Zygote16, 285–296 (2008). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

