Identifying intragenic functional modules of genomic variations associated with cancer phenotypes by learning representation of association networks
- PMID: 35794577
- PMCID: PMC9258200
- DOI: 10.1186/s12920-022-01298-6
Identifying intragenic functional modules of genomic variations associated with cancer phenotypes by learning representation of association networks
Abstract
Background: Genome-wide Association Studies (GWAS) aims to uncover the link between genomic variation and phenotype. They have been actively applied in cancer biology to investigate associations between variations and cancer phenotypes, such as susceptibility to certain types of cancer and predisposed responsiveness to specific treatments. Since GWAS primarily focuses on finding associations between individual genomic variations and cancer phenotypes, there are limitations in understanding the mechanisms by which cancer phenotypes are cooperatively affected by more than one genomic variation.
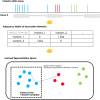
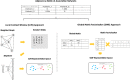
Results: This paper proposes a network representation learning approach to learn associations among genomic variations using a prostate cancer cohort. The learned associations are encoded into representations that can be used to identify functional modules of genomic variations within genes associated with early- and late-onset prostate cancer. The proposed method was applied to a prostate cancer cohort provided by the Veterans Administration's Million Veteran Program to identify candidates for functional modules associated with early-onset prostate cancer. The cohort included 33,159 prostate cancer patients, 3181 early-onset patients, and 29,978 late-onset patients. The reproducibility of the proposed approach clearly showed that the proposed approach can improve the model performance in terms of robustness.
Conclusions: To our knowledge, this is the first attempt to use a network representation learning approach to learn associations among genomic variations within genes. Associations learned in this way can lead to an understanding of the underlying mechanisms of how genomic variations cooperatively affect each cancer phenotype. This method can reveal unknown knowledge in the field of cancer biology and can be utilized to design more advanced cancer-targeted therapies.
Keywords: Genome-wide Association Study; Machine Learning; Network Representation Learning.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Glusman G, Rose PW, Prlić A, Dougherty J, Duarte JM, Hoffman AS, Barton GJ, Bendixen E, Bergquist T, Bock C, et al. Mapping genetic variations to three-dimensional protein structures to enhance variant interpretation: a proposed framework. Genome Med. 2017;9(1):1–10. doi: 10.1186/s13073-017-0509-y. - DOI - PMC - PubMed

