This is a preprint.
Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations
- PMID: 35794893
- PMCID: PMC9258294
- DOI: 10.21203/rs.3.rs-1723829/v1
Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations
Update in
-
Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations.Nat Methods. 2023 Apr;20(4):512-522. doi: 10.1038/s41592-023-01769-3. Epub 2023 Feb 23. Nat Methods. 2023. PMID: 36823332 Free PMC article.
Abstract
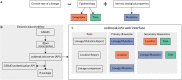
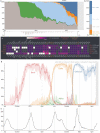
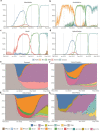
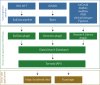
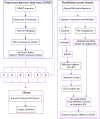
The emergence of SARS-CoV-2 variants of concern has prompted the need for near real-time genomic surveillance to inform public health interventions. In response to this need, the global scientific community, through unprecedented effort, has sequenced and shared over 11 million genomes through GISAID, as of May 2022. This extraordinarily high sampling rate provides a unique opportunity to track the evolution of the virus in near real-time. Here, we present outbreak.info, a platform that currently tracks over 40 million combinations of PANGO lineages and individual mutations, across over 7,000 locations, to provide insights for researchers, public health officials, and the general public. We describe the interpretable and opinionated visualizations in the variant and location focussed reports available in our web application, the pipelines that enable the scalable ingestion of heterogeneous sources of SARS-CoV-2 variant data, and the server infrastructure that enables widespread data dissemination via a high performance API that can be accessed using an R package. We present a case study that illustrates how outbreak.info can be used for genomic surveillance and as a hypothesis generation tool to understand the ongoing pandemic at varying geographic and temporal scales. With an emphasis on scalability, interactivity, interpretability, and reusability, outbreak.info provides a template to enable genomic surveillance at a global and localized scale.
Conflict of interest statement
Conflicts of Interest
MAS receives grants from the US National Institutes of Health within the scope of this work, and grants and contracts from the US Food & Drug Administration, the US Department of Veterans Affairs and Janssen Research & Development outside the scope of this work. MAS and KGA have received consulting fees and/or compensated expert testimony on SARS-CoV-2 and the COVID-19 pandemic.
Figures

References
-
- Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New England Journal of Medicine vol. 384 1576–1578 (2021). - PubMed
-
- edward_holmes et al. Novel 2019 coronavirus genome. https://virological.org/t/novel-2019-coronavirus-genome/319 (2020).
-
- GISAID - Initiative. https://gisaid.org.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

