This is a preprint.
Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection
- PMID: 35794895
- PMCID: PMC9258298
- DOI: 10.1101/2022.06.29.22276868
Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection
Update in
-
Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection.Cell Rep Med. 2023 Feb 21;4(2):100943. doi: 10.1016/j.xcrm.2023.100943. Epub 2023 Jan 27. Cell Rep Med. 2023. PMID: 36791724 Free PMC article.
Abstract
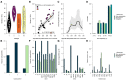
The chronic infection hypothesis for novel SARS-CoV-2 variant emergence is increasingly gaining credence following the appearance of Omicron. Here we investigate intrahost evolution and genetic diversity of lineage B.1.517 during a SARS-CoV-2 chronic infection lasting for 471 days (and still ongoing) with consistently recovered infectious virus and high viral loads. During the infection, we found an accelerated virus evolutionary rate translating to 35 nucleotide substitutions per year, approximately two-fold higher than the global SARS-CoV-2 evolutionary rate. This intrahost evolution led to the emergence and persistence of at least three genetically distinct genotypes suggesting the establishment of spatially structured viral populations continually reseeding different genotypes into the nasopharynx. Finally, using unique molecular indexes for accurate intrahost viral sequencing, we tracked the temporal dynamics of genetic diversity to identify advantageous mutations and highlight hallmark changes for chronic infection. Our findings demonstrate that untreated chronic infections accelerate SARS-CoV-2 evolution, ultimately providing opportunity for the emergence of genetically divergent and potentially highly transmissible variants as seen with Delta and Omicron.
Conflict of interest statement
Conflicts of interest
NDG is a consultant for Tempus Labs and the National Basketball Association for work related to COVID-19 but is outside the submitted work. UNC is pursuing intellectual property protection for Primer ID sequencing and RS has received nominal royalties from licensing.
Figures





References
-
- Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. (2020). Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell 183, 1901–1912.e9.. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
