Curation of a reference database of COI sequences for insect identification through DNA metabarcoding: COins
- PMID: 35796594
- PMCID: PMC9261288
- DOI: 10.1093/database/baac055
Curation of a reference database of COI sequences for insect identification through DNA metabarcoding: COins
Abstract
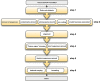
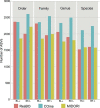
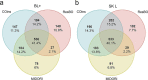
DNA metabarcoding is a widespread approach for the molecular identification of organisms. While the associated wet-lab and data processing procedures are well established and highly efficient, the reference databases for taxonomic assignment can be implemented to improve the accuracy of identifications. Insects are among the organisms for which DNA-based identification is most commonly used; yet, a DNA-metabarcoding reference database specifically curated for their species identification using software requiring local databases is lacking. Here, we present COins, a database of 5' region cytochrome c oxidase subunit I sequences (COI-5P) of insects that includes over 532 000 representative sequences of >106 000 species specifically formatted for the QIIME2 software platform. Through a combination of automated and manually curated steps, we developed this database starting from all COI sequences available in the Barcode of Life Data System for insects, focusing on sequences that comply with several standards, including a species-level identification. COins was validated on previously published DNA-metabarcoding sequences data (bulk samples from Malaise traps) and its efficiency compared with other publicly available reference databases (not specific for insects). COins can allow an increase of up to 30% of species-level identifications and thus can represent a valuable resource for the taxonomic assignment of insects' DNA-metabarcoding data, especially when species-level identification is needed https://doi.org/10.6084/m9.figshare.19130465.v1.
© The Author(s) 2022. Published by Oxford University Press.
Figures