CRISPR-Cas9-mediated mutagenesis of kiwifruit BFT genes results in an evergrowing but not early flowering phenotype
- PMID: 35796629
- PMCID: PMC9616528
- DOI: 10.1111/pbi.13888
CRISPR-Cas9-mediated mutagenesis of kiwifruit BFT genes results in an evergrowing but not early flowering phenotype
Abstract
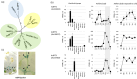
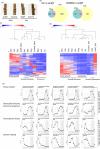
Phosphatidylethanolamine-binding protein (PEBP) genes regulate flowering and architecture in many plant species. Here, we study kiwifruit (Actinidia chinensis, Ac) PEBP genes with homology to BROTHER OF FT AND TFL1 (BFT). CRISPR-Cas9 was used to target AcBFT genes in wild-type and fast-flowering kiwifruit backgrounds. The editing construct was designed to preferentially target AcBFT2, whose expression is elevated in dormant buds. Acbft lines displayed an evergrowing phenotype and increased branching, while control plants established winter dormancy. The evergrowing phenotype, encompassing delayed budset and advanced budbreak after defoliation, was identified in multiple independent lines with edits in both alleles of AcBFT2. RNA-seq analyses conducted using buds from gene-edited and control lines indicated that Acbft evergrowing plants had a transcriptome similar to that of actively growing wild-type plants, rather than dormant controls. Mutations in both alleles of AcBFT2 did not promote flowering in wild-type or affect flowering time, morphology and fertility in fast-flowering transgenic kiwifruit. In summary, editing of AcBFT2 has the potential to reduce plant dormancy with no adverse effect on flowering, giving rise to cultivars better suited for a changing climate.
Keywords: Actinidia chinensis; BFT; CRISPR-Cas9; dormancy; flowering; kiwifruit.
© 2022 The New Zealand Institute for Plant and Food Research Ltd and The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Abelenda, J.A. , Bergonzi, S. , Oortwijn, M. , Sonnewald, S. , Du, M. , Visser, R.G.F. , Sonnewald, U. et al. (2019) Source‐sink regulation is mediated by interaction of an FT homolog with a SWEET protein in potato. Curr. Biol. 29, 1178–1186.e1176. - PubMed
-
- Akagi, T. , Pilkington, S.M. , Varkonyi‐Gasic, E. , Henry, I.M. , Sugano, S.S. , Sonoda, M. , Firl, A. et al. (2019) Two Y‐chromosome‐encoded genes determine sex in kiwifruit. Nat. Plants 5, 801–809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

