Interaction between endogenous microRNAs and virus-derived small RNAs controls viral replication in insect vectors
- PMID: 35797383
- PMCID: PMC9295959
- DOI: 10.1371/journal.ppat.1010709
Interaction between endogenous microRNAs and virus-derived small RNAs controls viral replication in insect vectors
Abstract
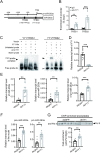
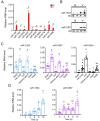
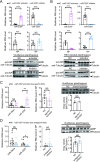
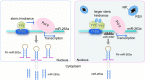
MicroRNAs (miRNAs) play an important role in resisting virus infection in insects. Viruses are recognized by insect RNA interference systems, which generate virus-derived small RNAs (vsRNAs). To date, it is unclear whether viruses employ vsRNAs to regulate the expression of endogenous miRNAs. We previously found that miR-263a facilitated the proliferation of rice stripe virus (RSV) in the insect vector small brown planthopper. However, miR-263a was significantly downregulated by RSV. Here, we deciphered the regulatory mechanisms of RSV on miR-263a expression. The promoter region of miR-263a was characterized, and the transcription factor YY1 was found to negatively regulate the transcription of miR-263a. The nucleocapsid protein of RSV promoted the inhibitory effect of YY1 on miR-263a transcription by reducing the binding ability of RNA polymerase II to the promoter of miR-263a. Moreover, an RSV-derived small RNA, vsR-3397, downregulated miR-263a transcription by directly targeting the promoter region with partial sequence complementarity. The reduction in miR-263a suppressed RSV replication and was beneficial for maintaining a tolerable accumulation level of RSV in insect vectors. This dual regulation mechanism reflects an ingenious adaptation strategy of viruses to their insect vectors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
A double-agent microRNA regulates viral cross-kingdom infection in animals and plants.EMBO J. 2025 May;44(9):2446-2472. doi: 10.1038/s44318-025-00405-4. Epub 2025 Mar 5. EMBO J. 2025. PMID: 40045022 Free PMC article.
-
Coordination between terminal variation of the viral genome and insect microRNAs regulates rice stripe virus replication in insect vectors.PLoS Pathog. 2021 Mar 10;17(3):e1009424. doi: 10.1371/journal.ppat.1009424. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33690727 Free PMC article.
-
Alternative Splicing Landscape of Small Brown Planthopper and Different Response of JNK2 Isoforms to Rice Stripe Virus Infection.J Virol. 2022 Jan 26;96(2):e0171521. doi: 10.1128/JVI.01715-21. Epub 2021 Nov 10. J Virol. 2022. PMID: 34757837 Free PMC article.
-
Ribosomal protein L18 is an essential factor that promote rice stripe virus accumulation in small brown planthopper.Virus Res. 2018 Mar 2;247:15-20. doi: 10.1016/j.virusres.2018.01.011. Epub 2018 Jan 31. Virus Res. 2018. PMID: 29374519
-
The small brown planthopper (Laodelphax striatellus) as a vector of the rice stripe virus.Arch Insect Biochem Physiol. 2023 Feb;112(2):e21992. doi: 10.1002/arch.21992. Epub 2022 Dec 27. Arch Insect Biochem Physiol. 2023. PMID: 36575628 Review.
Cited by
-
A plant virus manipulates the long-winged morph of insect vectors.Proc Natl Acad Sci U S A. 2024 Jan 16;121(3):e2315341121. doi: 10.1073/pnas.2315341121. Epub 2024 Jan 8. Proc Natl Acad Sci U S A. 2024. PMID: 38190519 Free PMC article.
-
Small interfering RNAs generated from the terminal panhandle structure of negative-strand RNA virus promote viral infection.PLoS Pathog. 2025 Jan 3;21(1):e1012789. doi: 10.1371/journal.ppat.1012789. eCollection 2025 Jan. PLoS Pathog. 2025. PMID: 39752360 Free PMC article.
-
A Review of Vector-Borne Rice Viruses.Viruses. 2022 Oct 14;14(10):2258. doi: 10.3390/v14102258. Viruses. 2022. PMID: 36298813 Free PMC article. Review.
-
Host cells reprogram lipid droplet synthesis through YY1 to resist PRRSV infection.mBio. 2024 Aug 14;15(8):e0154924. doi: 10.1128/mbio.01549-24. Epub 2024 Jul 2. mBio. 2024. PMID: 38953350 Free PMC article.
-
A double-agent microRNA regulates viral cross-kingdom infection in animals and plants.EMBO J. 2025 May;44(9):2446-2472. doi: 10.1038/s44318-025-00405-4. Epub 2025 Mar 5. EMBO J. 2025. PMID: 40045022 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

