Unsupervised inter-frame motion correction for whole-body dynamic PET using convolutional long short-term memory in a convolutional neural network
- PMID: 35797734
- PMCID: PMC10923189
- DOI: 10.1016/j.media.2022.102524
Unsupervised inter-frame motion correction for whole-body dynamic PET using convolutional long short-term memory in a convolutional neural network
Abstract
Subject motion in whole-body dynamic PET introduces inter-frame mismatch and seriously impacts parametric imaging. Traditional non-rigid registration methods are generally computationally intense and time-consuming. Deep learning approaches are promising in achieving high accuracy with fast speed, but have yet been investigated with consideration for tracer distribution changes or in the whole-body scope. In this work, we developed an unsupervised automatic deep learning-based framework to correct inter-frame body motion. The motion estimation network is a convolutional neural network with a combined convolutional long short-term memory layer, fully utilizing dynamic temporal features and spatial information. Our dataset contains 27 subjects each under a 90-min FDG whole-body dynamic PET scan. Evaluating performance in motion simulation studies and a 9-fold cross-validation on the human subject dataset, compared with both traditional and deep learning baselines, we demonstrated that the proposed network achieved the lowest motion prediction error, obtained superior performance in enhanced qualitative and quantitative spatial alignment between parametric Ki and Vb images, and significantly reduced parametric fitting error. We also showed the potential of the proposed motion correction method for impacting downstream analysis of the estimated parametric images, improving the ability to distinguish malignant from benign hypermetabolic regions of interest. Once trained, the motion estimation inference time of our proposed network was around 460 times faster than the conventional registration baseline, showing its potential to be easily applied in clinical settings.
Keywords: Convolutional network; Long-short term memory; Motion correction; Parametric imaging; Whole-body dynamic PET.
Copyright © 2022. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest David Pigg, Bruce Spottiswoode and Michael E. Casey are employees of Siemens Medical Solutions USA, Inc. No other potential conflicts of interest relevant to this article exist.
Figures
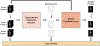
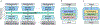



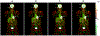
References
-
- Abdi AH, Luong C, Tsang T, Jue J, Gin K, Yeung D, Hawley D, Rohling R, Abol-maesumi P, 2017. Quality assessment of echocardiographic cine using recurrent neural networks: Feasibility on five standard view planes. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 302–310.
-
- Arbelle A, Raviv TR, 2019. Microscopy cell segmentation via convolutional lstm networks. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). IEEE, pp. 1008–1012.
-
- Azad R, Asadi-Aghbolaghi M, Fathy M, Escalera S, 2019. Bi-directional convlstm u-net with densley connected convolutions. In: Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops. 0–0
-
- Azizmohammadi F, Martin R, Miro J, Duong L, 2019. Model-free cardiorespiratory motion prediction from x-ray angiography sequence with lstm network. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE, pp. 7014–7018. - PubMed
-
- Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV, 2019. Voxelmorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38 (8), 1788–1800. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

