Characterization of SARS-CoV-2 Spike mutations important for infection of mice and escape from human immune sera
- PMID: 35798721
- PMCID: PMC9261898
- DOI: 10.1038/s41467-022-30763-0
Characterization of SARS-CoV-2 Spike mutations important for infection of mice and escape from human immune sera
Abstract
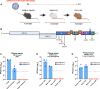
Due to differences in human and murine angiotensin converting enzyme 2 (ACE-2) receptor, initially available SARS-CoV-2 isolates could not infect mice. Here we show that serial passaging of USA-WA1/2020 strain in mouse lungs results in "mouse-adapted" SARS-CoV-2 (MA-SARS-CoV-2) with mutations in S, M, and N genes, and a twelve-nucleotide insertion in the S gene. MA-SARS-CoV-2 infection causes mild disease, with more pronounced morbidity depending on genetic background and in aged and obese mice. Two mutations in the S gene associated with mouse adaptation (N501Y, H655Y) are present in SARS-CoV-2 variants of concern (VoCs). N501Y in the receptor binding domain of viruses of the B.1.1.7, B.1.351, P.1 and B.1.1.529 lineages (Alpha, Beta, Gamma and Omicron variants) is associated with high transmissibility and allows VoCs to infect wild type mice. We further show that S protein mutations of MA-SARS-CoV-2 do not affect neutralization efficiency by human convalescent and post vaccination sera.
© 2022. The Author(s).
Conflict of interest statement
The A.G.-S. laboratory has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, ImmunityBio, and Nanocomposix. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, and Esperovax. M.S. has licensed SARS-CoV-2 serological assays to commercial entities and has filed for patent protection for serological assays as well as SARS-CoV-2 vaccines. F.K. is listed as inventors on the pending patent applications. The F.K. laboratory has received research support from GSK, Dynavax, and Pfizer. F.K. has in the past received consulting fees from Curevac, Merck, Pfizer, and Seqirus. The J.A.R. laboratory received support from Tonix Pharmaceuticals, Xing Technologies and Zoetis, outside of the reported work. J.A.R. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections, owned by Kansas State University, KS, or the Icahn School of Medicine at Mount Sinai, New York. A provisional patent application on a “A novel 4 Amino Acid Insertion into the Spike Protein of SARS-CoV-2” was submitted by KSU in July 2020 with C.D.M., D.A.M., and J.A.R. listed as inventors. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

