Characterization of regulatory transcriptional mechanisms in hepatocyte lipotoxicity
- PMID: 35798791
- PMCID: PMC9262951
- DOI: 10.1038/s41598-022-15731-4
Characterization of regulatory transcriptional mechanisms in hepatocyte lipotoxicity
Abstract
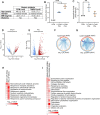
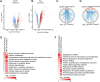
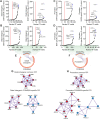
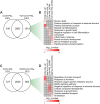
Non-alcoholic fatty liver disease is a continuum of disorders among which non-alcoholic steatohepatitis (NASH) is particularly associated with a negative prognosis. Hepatocyte lipotoxicity is one of the main pathogenic factors of liver fibrosis and NASH. However, the molecular mechanisms regulating this process are poorly understood. The main aim of this study was to dissect transcriptional mechanisms regulated by lipotoxicity in hepatocytes. We achieved this aim by combining transcriptomic, proteomic and chromatin accessibility analyses from human liver and mouse hepatocytes. This integrative approach revealed several transcription factor networks deregulated by NASH and lipotoxicity. To validate these predictions, genetic deletion of the transcription factors MAFK and TCF4 was performed, resulting in hepatocytes that were better protected against saturated fatty acid oversupply. MAFK- and TCF4-regulated gene expression profiles suggest a mitigating effect against cell stress, while promoting cell survival and growth. Moreover, in the context of lipotoxicity, some MAFK and TCF4 target genes were to the corresponding differentially regulated transcripts in human liver fibrosis. Collectively, our findings comprehensively profile the transcriptional response to lipotoxicity in hepatocytes, revealing new molecular insights and providing a valuable resource for future endeavours to tackle the molecular mechanisms of NASH.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

