Enhancing untargeted metabolomics using metadata-based source annotation
- PMID: 35798960
- PMCID: PMC10277029
- DOI: 10.1038/s41587-022-01368-1
Enhancing untargeted metabolomics using metadata-based source annotation
Erratum in
-
Author Correction: Enhancing untargeted metabolomics using metadata-based source annotation.Nat Biotechnol. 2023 Nov;41(11):1656. doi: 10.1038/s41587-023-02025-x. Nat Biotechnol. 2023. PMID: 37853256 No abstract available.
Abstract
Human untargeted metabolomics studies annotate only ~10% of molecular features. We introduce reference-data-driven analysis to match metabolomics tandem mass spectrometry (MS/MS) data against metadata-annotated source data as a pseudo-MS/MS reference library. Applying this approach to food source data, we show that it increases MS/MS spectral usage 5.1-fold over conventional structural MS/MS library matches and allows empirical assessment of dietary patterns from untargeted data.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures
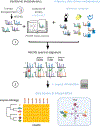
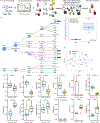
References
Publication types
MeSH terms
Grants and funding
- R01 AG046171/AG/NIA NIH HHS/United States
- R01 MD011389/MD/NIMHD NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- R01 AG061066/AG/NIA NIH HHS/United States
- UL1 TR002535/TR/NCATS NIH HHS/United States
- K12 HD000850/HD/NICHD NIH HHS/United States
- T32 DK007202/DK/NIDDK NIH HHS/United States
- U01 AG061359/AG/NIA NIH HHS/United States
- P50 AR060772/AR/NIAMS NIH HHS/United States
- RF1 AG058942/AG/NIA NIH HHS/United States
- T32 HL149646/HL/NHLBI NIH HHS/United States
- K12 GM068524/GM/NIGMS NIH HHS/United States
- T32 AR064194/AR/NIAMS NIH HHS/United States
- DP1 AT010885/AT/NCCIH NIH HHS/United States
- U19 AG063744/AG/NIA NIH HHS/United States
- P30 DK120515/DK/NIDDK NIH HHS/United States
- R21 AR075990/AR/NIAMS NIH HHS/United States
- DP2 AT010401/AT/NCCIH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

