A Novel Angiogenesis-Related Gene Signature to Predict Biochemical Recurrence of Patients with Prostate Cancer following Radical Therapy
- PMID: 35799610
- PMCID: PMC9256390
- DOI: 10.1155/2022/2448428
A Novel Angiogenesis-Related Gene Signature to Predict Biochemical Recurrence of Patients with Prostate Cancer following Radical Therapy
Abstract
Background: Prostate cancer (PCa) is one of the most common malignancies in males; we aim to establish a novel angiogenesis-related gene signature for biochemical recurrence (BCR) prediction in PCa patients following radical therapy.
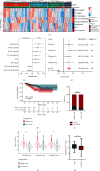
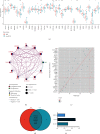
Methods: Gene expression profiles and corresponding clinicopathological data were acquired from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) database. We quantified the levels of various cancer hallmarks and identified angiogenesis as the primary risk factor for BCR. Then machine learning methods combined with Cox regression analysis were used to screen prognostic genes and construct an angiogenesis-related gene signature, which was further verified in external cohorts. Furthermore, estimation of immune cell abundance and prediction of drug responses were also conducted to detect potential mechanisms.
Results: Angiogenesis was regarded as the leading risk factor for BCR in PCa patients (HR = 1.58, 95% CI: 1.38-1.81), and a novel prognostic signature based on three genes (NRP1, JAG2, and VCAN) was developed in the training cohort and successfully validated in another three independent cohorts. In all datasets, this signature was identified as a prognostic factor with promising and robust predictive values. Moreover, it also predicted higher infiltration of regulatory T cells and M2-polarized macrophages and increased drug sensitivity of angiogenesis inhibitors in high-risk patients.
Conclusions: The angiogenesis-related three-gene signature may serve as an independent prognostic factor for BCR, which would contribute to risk stratification and personalized management of PCa patients after radical therapy in clinical practice.
Copyright © 2022 Bohan Fan et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



Similar articles
-
A Novel Set of Immune-associated Gene Signature predicts Biochemical Recurrence in Localized Prostate Cancer Patients after Radical Prostatectomy.J Cancer. 2021 May 1;12(12):3715-3725. doi: 10.7150/jca.51059. eCollection 2021. J Cancer. 2021. PMID: 33995646 Free PMC article.
-
Identification and validation of a six immune-related gene signature for prediction of biochemical recurrence in localized prostate cancer following radical prostatectomy.Transl Androl Urol. 2021 Mar;10(3):1018-1029. doi: 10.21037/tau-20-1231. Transl Androl Urol. 2021. PMID: 33850736 Free PMC article.
-
A Novel Apoptosis-Related Gene Signature Predicts Biochemical Recurrence of Localized Prostate Cancer After Radical Prostatectomy.Front Genet. 2020 Nov 30;11:586376. doi: 10.3389/fgene.2020.586376. eCollection 2020. Front Genet. 2020. PMID: 33329725 Free PMC article.
-
A new risk stratification system of prostate cancer to identify high-risk biochemical recurrence patients.Transl Androl Urol. 2020 Dec;9(6):2572-2586. doi: 10.21037/tau-20-1019. Transl Androl Urol. 2020. PMID: 33457230 Free PMC article.
-
A novel gene signature to predict immune infiltration and outcome in patients with prostate cancer.Oncoimmunology. 2020 Jun 1;9(1):1762473. doi: 10.1080/2162402X.2020.1762473. Oncoimmunology. 2020. PMID: 32923125 Free PMC article.
Cited by
-
Predicting prostate cancer recurrence: Introducing PCRPS, an advanced online web server.Heliyon. 2024 Mar 29;10(7):e28878. doi: 10.1016/j.heliyon.2024.e28878. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38623253 Free PMC article.
-
Comparative efficacy of radical prostatectomy and radiotherapy in the treatment of high-risk prostate cancer.Technol Health Care. 2024;32(6):4671-4679. doi: 10.3233/THC-240910. Technol Health Care. 2024. PMID: 39093097 Free PMC article.
References
-
- Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2020. CA: A Cancer Journal For Clinicians . 2020;70 - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

