Diverse Aquatic Animal Matrices Play a Key Role in Survival and Potential Virulence of Non-O1/O139 Vibrio cholerae Isolates
- PMID: 35801116
- PMCID: PMC9255913
- DOI: 10.3389/fmicb.2022.896767
Diverse Aquatic Animal Matrices Play a Key Role in Survival and Potential Virulence of Non-O1/O139 Vibrio cholerae Isolates
Abstract
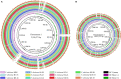
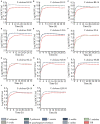
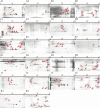
Vibrio cholerae can cause pandemic cholera in humans. The waterborne bacterium is frequently isolated from aquatic products worldwide. However, current literature on the impact of aquatic product matrices on the survival and pathogenicity of cholerae is rare. In this study, the growth of eleven non-O1/0O139 V. cholerae isolates recovered from eight species of commonly consumed fish and shellfish was for the first time determined in the eight aquatic animal matrices, most of which highly increased the bacterial biomass when compared with routine trypsin soybean broth (TSB) medium. Secretomes of the V. cholerae isolates (draft genome size: 3,852,021-4,144,013 bp) were determined using two-dimensional gel electrophoresis (2DE-GE) and liquid chromatography-tandem mass spectrometry (LC-MS/MS) techniques. Comparative secretomic analyses revealed 74 differential extracellular proteins, including several virulence- and resistance-associated proteins secreted by the V. cholerae isolates when grown in the eight matrices. Meanwhile, a total of 8,119 intracellular proteins were identified, including 83 virulence- and 8 resistance-associated proteins, of which 61 virulence-associated proteins were absent from proteomes of these isolates when grown in the TSB medium. Additionally, comparative genomic and proteomic analyses also revealed several strain-specific proteins with unknown functions in the V. cholerae isolates. Taken, the results in this study demonstrate that distinct secretomes and proteomes induced by the aquatic animal matrices facilitate V. cholerae resistance in the edible aquatic animals and enhance the pathogenicity of the leading waterborne pathogen worldwide.
Keywords: V. cholerae; aquatic product matrix; genome; proteome; resistance; secretome; virulence.
Copyright © 2022 Yan, Jin, Zhang, Xu, Peng, Qin and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
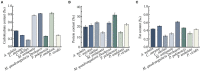
References
LinkOut - more resources
Full Text Sources

