Characterization of the complete chloroplast genome of the medicinal herb Veronica polita Fr. 1819 (Lamiales: Plantaginaceae)
- PMID: 35801139
- PMCID: PMC9255118
- DOI: 10.1080/23802359.2022.2086074
Characterization of the complete chloroplast genome of the medicinal herb Veronica polita Fr. 1819 (Lamiales: Plantaginaceae)
Abstract
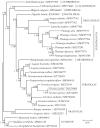
Veronica polita Fr. 1819 (synonym: Veronica didyma Ten. 1981) is a species of annual herb with high medicinal value. It is originally from Southwest Asia, but has been naturalized widely in many regions of the world. In this study, the complete chloroplast genome of V. polita was determined to be 150,191 bp long with a typical quadripartite structure, comprising two inverted repeat regions (IRa and IRb, 25,465 bp each), a large single-copy (LSC) region (81,847 bp) and a small single-copy (SSC) region (17,414 bp). It encodes a panel of 114 genes (including 79 protein-coding, 31 tRNA, and four rRNA genes) with 18 of them being completely or partially duplicated and 19 of them possessing one or two introns. Phylogenetic analysis supported the tribal-level taxonomy of the family Plantaginaceae, and revealed that V. polita was most closely related to the congener Veronica persica Poir. 1808.
Keywords: Chloroplast genome; Veronica polita Fr. 1819; high-throughput sequencing; medicinal herb; phylogenetic analysis.
© 2022 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
The authors report no conflict of interest.
Figures
Similar articles
-
Characterization of the complete chloroplast genome of Veronica arvensis and its phylogenomic inference in plantaginaceae.Mitochondrial DNA B Resour. 2022 Nov 4;7(11):1928-1932. doi: 10.1080/23802359.2022.2139162. eCollection 2022. Mitochondrial DNA B Resour. 2022. PMID: 36353054 Free PMC article.
-
Characterization of the complete plastid genome of of Veronica eriogyne H. Winkl., a Tibetan medicinal herb.Mitochondrial DNA B Resour. 2021 Nov 12;6(12):3402-3403. doi: 10.1080/23802359.2021.1998802. eCollection 2021. Mitochondrial DNA B Resour. 2021. PMID: 34790875 Free PMC article.
-
Characterization of the complete chloroplast genome sequence of Limnophila sessiliflora Blume 1826 (Plantaginaceae).Mitochondrial DNA B Resour. 2023 May 18;8(5):585-588. doi: 10.1080/23802359.2023.2209390. eCollection 2023. Mitochondrial DNA B Resour. 2023. PMID: 37213788 Free PMC article.
-
The complete chloroplast genome of Hippuris vulgaris (Plantaginaceae).Mitochondrial DNA B Resour. 2021 Jan 28;6(1):259-260. doi: 10.1080/23802359.2020.1860706. Mitochondrial DNA B Resour. 2021. PMID: 33553638 Free PMC article.
-
Characterization and phylogenetic analysis of the complete plastome of Veronica undulata (Plantaginaceae).Mitochondrial DNA B Resour. 2021 Aug 19;6(9):2706-2707. doi: 10.1080/23802359.2021.1966345. eCollection 2021. Mitochondrial DNA B Resour. 2021. PMID: 34435127 Free PMC article.
Cited by
-
The chloroplast genomes of two medicinal species (Veronica anagallis-aquatica L. and Veronica undulata Wall.) and its comparative analysis with related Veronica species.Sci Rep. 2024 Jun 17;14(1):13945. doi: 10.1038/s41598-024-64896-7. Sci Rep. 2024. PMID: 38886540 Free PMC article.
References
-
- Akanda MR, Nam H-H, Tian W, Islam A, Choo B-K, Park B-Y.. 2018. Regulation of JAK2/STAT3 and NF-κB signal transduction pathways; Veronica polita alleviates dextran sulfate sodium-induced murine colitis. Biomed Pharmacother. 100:296–303. - PubMed
-
- Fowler RM, McLay TGB, Schuster TM, Buirchell BJ, Murphy DJ, Bayly MJ.. 2020. Plastid phylogenomic analysis of tribe Myoporeae (Scrophulariaceae). Plant Syst Evol. 306(3):52.
LinkOut - more resources
Full Text Sources
