Validation of a multi-ancestry polygenic risk score and age-specific risks of prostate cancer: A meta-analysis within diverse populations
- PMID: 35801699
- PMCID: PMC9322982
- DOI: 10.7554/eLife.78304
Validation of a multi-ancestry polygenic risk score and age-specific risks of prostate cancer: A meta-analysis within diverse populations
Abstract
Background: We recently developed a multi-ancestry polygenic risk score (PRS) that effectively stratifies prostate cancer risk across populations. In this study, we validated the performance of the PRS in the multi-ancestry Million Veteran Program and additional independent studies.
Methods: Within each ancestry population, the association of PRS with prostate cancer risk was evaluated separately in each case-control study and then combined in a fixed-effects inverse-variance-weighted meta-analysis. We further assessed the effect modification by age and estimated the age-specific absolute risk of prostate cancer for each ancestry population.
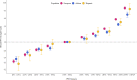
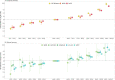
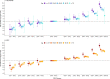
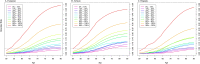
Results: The PRS was evaluated in 31,925 cases and 490,507 controls, including men from European (22,049 cases, 414,249 controls), African (8794 cases, 55,657 controls), and Hispanic (1082 cases, 20,601 controls) populations. Comparing men in the top decile (90-100% of the PRS) to the average 40-60% PRS category, the prostate cancer odds ratio (OR) was 3.8-fold in European ancestry men (95% CI = 3.62-3.96), 2.8-fold in African ancestry men (95% CI = 2.59-3.03), and 3.2-fold in Hispanic men (95% CI = 2.64-3.92). The PRS did not discriminate risk of aggressive versus nonaggressive prostate cancer. However, the OR diminished with advancing age (European ancestry men in the top decile: ≤55 years, OR = 7.11; 55-60 years, OR = 4.26; >70 years, OR = 2.79). Men in the top PRS decile reached 5% absolute prostate cancer risk ~10 years younger than men in the 40-60% PRS category.
Conclusions: Our findings validate the multi-ancestry PRS as an effective prostate cancer risk stratification tool across populations. A clinical study of PRS is warranted to determine whether the PRS could be used for risk-stratified screening and early detection.
Funding: This work was supported by the National Cancer Institute at the National Institutes of Health (grant numbers U19 CA214253 to C.A.H., U01 CA257328 to C.A.H., U19 CA148537 to C.A.H., R01 CA165862 to C.A.H., K99 CA246063 to B.F.D, and T32CA229110 to F.C), the Prostate Cancer Foundation (grants 21YOUN11 to B.F.D. and 20CHAS03 to C.A.H.), the Achievement Rewards for College Scientists Foundation Los Angeles Founder Chapter to B.F.D, and the Million Veteran Program-MVP017. This research has been conducted using the UK Biobank Resource under application number 42195. This research is based on data from the Million Veteran Program, Office of Research and Development, and the Veterans Health Administration. This publication does not represent the views of the Department of Veteran Affairs or the United States Government.
Keywords: African ancestry; Hispanic; epidemiology; genetics; genomics; global health; health disparities; multi-ancestry; none; polygenic risk scores; prostate cancer.
Conflict of interest statement
FC, AR, XS, CR, CA, WT, AK, AP, KC, MJ, SG, LN, OO, OP, AA, OA, HA, MJ, OO, MN, BA, SM, AD, JM, AA, HD, JL, TR, SA, JG, AJ, DC, CH No competing interests declared, BD received honorarium for presentations at Society of Urology Oncology Annual Meeting (2021) and the Social Genomics Group at the University of Wisconsin, Madison (2021). The author has no other competing interests to declare, RM has stock or stock options in Navipoint Genomics LLC. The author has no other competing interests to declare
Figures

References
-
- Amin Al Olama A, Benlloch S, Antoniou AC, Giles GG, Severi G, Neal DE, Hamdy FC, Donovan JL, Muir K, Schleutker J, Henderson BE, Haiman CA, Schumacher FR, Pashayan N, Pharoah PDP, Ostrander EA, Stanford JL, Batra J, Clements JA, Chambers SK, Weischer M, Nordestgaard BG, Ingles SA, Sorensen KD, Orntoft TF, Park JY, Cybulski C, Maier C, Doerk T, Dickinson JL, Cannon-Albright L, Brenner H, Rebbeck TR, Zeigler-Johnson C, Habuchi T, Thibodeau SN, Cooney KA, Chappuis PO, Hutter P, Kaneva RP, Foulkes WD, Zeegers MP, Lu YJ, Zhang HW, Stephenson R, Cox A, Southey MC, Spurdle AB, FitzGerald L, Leongamornlert D, Saunders E, Tymrakiewicz M, Guy M, Dadaev T, Little SJ, Govindasami K, Sawyer E, Wilkinson R, Herkommer K, Hopper JL, Lophatonanon A, Rinckleb AE, Kote-Jarai Z, Eeles RA, Easton DF, UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology. UK ProtecT Study Collaborators. PRACTICAL Consortium Risk analysis of prostate cancer in PRACTICAL, a multinational consortium, using 25 known prostate cancer susceptibility loci. Cancer Epidemiology, Biomarkers & Prevention. 2015;24:1121–1129. doi: 10.1158/1055-9965.EPI-14-0317. - DOI - PMC - PubMed
-
- Andrews C, Fortier B, Hayward A, Lederman R, Petersen L, McBride J, Petersen DC, Ajayi O, Kachambwa P, Seutloali M, Shoko A, Mokhosi M, Hiller R, Adams M, Ongaco C, Pugh E, Romm J, Shelford T, Chinegwundoh F, Adusei B, Mante S, Snyper NY, Agalliu I, Lounsbury DW, Rohan T, Orfanos A, Quintana Y, Jacobson JS, Neugut AI, Gelmann E, Lachance J, Dial C, Diallo TA, Jalloh M, Gueye SM, Kane PMS, Diop H, Ndiaye AJ, Sall AS, Toure-Kane NC, Onyemata E, Abimiku A, Adjei AA, Biritwum R, Gyasi R, Kyei M, Mensah JE, Okine J, Okyne V, Rockson I, Tay E, Tettey Y, Yeboah E, Chen WC, Singh E, Cook MB, Duffy CN, Hsing A, Soo CC, Fernandez P, Irusen H, Aisuodionoe-Shadrach O, Jamda AM, Olabode PO, Nwegbu MM, Ajibola OH, Ajamu OJ, Ambuwa YG, Adebiyi AO, Asuzu M, Ogunbiyi O, Popoola O, Shittu O, Amodu O, Odiaka E, Makinde I, Joffe M, Pentz A, Rebbeck TR. Development, evaluation, and implementation of a pan-african cancer research network: Men of african descent and carcinoma of the prostate. Journal of Global Oncology. 2018;4:1–14. doi: 10.1200/JGO.18.00063. - DOI - PMC - PubMed
-
- Antoniou AC, Beesley J, McGuffog L, Sinilnikova OM, Healey S, Neuhausen SL, Ding YC, Rebbeck TR, Weitzel JN, Lynch HT, Isaacs C, Ganz PA, Tomlinson G, Olopade OI, Couch FJ, Wang X, Lindor NM, Pankratz VS, Radice P, Manoukian S, Peissel B, Zaffaroni D, Barile M, Viel A, Allavena A, Dall’Olio V, Peterlongo P, Szabo CI, Zikan M, Claes K, Poppe B, Foretova L, Mai PL, Greene MH, Rennert G, Lejbkowicz F, Glendon G, Ozcelik H, Andrulis IL, Thomassen M, Gerdes AM, Sunde L, Cruger D, Birk Jensen U, Caligo M, Friedman E, Kaufman B, Laitman Y, Milgrom R, Dubrovsky M, Cohen S, Borg A, Jernström H, Lindblom A, Rantala J, Stenmark-Askmalm M, Melin B, Nathanson K, Domchek S, Jakubowska A, Lubinski J, Huzarski T, Osorio A, Lasa A, Durán M, Tejada MI, Godino J, Benitez J, Hamann U, Kriege M, Hoogerbrugge N, van der Luijt RB, van Asperen CJ, Devilee P, Meijers-Heijboer EJ, Blok MJ, Aalfs CM, Hogervorst F, Rookus M, Cook M, Oliver C, Frost D, Conroy D, Evans DG, Lalloo F, Pichert G, Davidson R, Cole T, Cook J, Paterson J, Hodgson S, Morrison PJ, Porteous ME, Walker L, Kennedy MJ, Dorkins H, Peock S, Godwin AK, Stoppa-Lyonnet D, de Pauw A, Mazoyer S, Bonadona V, Lasset C, Dreyfus H, Leroux D, Hardouin A, Berthet P, Faivre L, Loustalot C, Noguchi T, Sobol H, Rouleau E, Nogues C, Frénay M, Vénat-Bouvet L, Hopper JL, Daly MB, Terry MB, John EM, Buys SS, Yassin Y, Miron A, Goldgar D, Singer CF, Dressler AC, Gschwantler-Kaulich D, Pfeiler G, Hansen TVO, Jønson L, Agnarsson BA, Kirchhoff T, Offit K, Devlin V, Dutra-Clarke A, Piedmonte M, Rodriguez GC, Wakeley K, Boggess JF, Basil J, Schwartz PE, Blank SV, Toland AE, Montagna M, Casella C, Imyanitov E, Tihomirova L, Blanco I, Lazaro C, Ramus SJ, Sucheston L, Karlan BY, Gross J, Schmutzler R, Wappenschmidt B, Engel C, Meindl A, Lochmann M, Arnold N, Heidemann S, Varon-Mateeva R, Niederacher D, Sutter C, Deissler H, Gadzicki D, Preisler-Adams S, Kast K, Schönbuchner I, Caldes T, de la Hoya M, Aittomäki K, Nevanlinna H, Simard J, Spurdle AB, Holland H, Chen X, Platte R, Chenevix-Trench G, Easton DF, Ontario Cancer Genetics Network. SWE-BRCA. HEBON. EMBRACE. GEMO. Breast Cancer Family Registry. kConFab. CIMBA Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Research. 2010;70:9742–9754. doi: 10.1158/0008-5472.CAN-10-1907. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

