Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease
- PMID: 35801916
- PMCID: PMC9371922
- DOI: 10.1093/nar/gkac589
Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease
Abstract
Coronaviruses generate double-stranded (ds) RNA intermediates during viral replication that can activate host immune sensors. To evade activation of the host pattern recognition receptor MDA5, coronaviruses employ Nsp15, which is a uridine-specific endoribonuclease. Nsp15 is proposed to associate with the coronavirus replication-transcription complex within double-membrane vesicles to cleave these dsRNA intermediates. How Nsp15 recognizes and processes dsRNA is poorly understood because previous structural studies of Nsp15 have been limited to small single-stranded (ss) RNA substrates. Here we present cryo-EM structures of SARS-CoV-2 Nsp15 bound to a 52nt dsRNA. We observed that the Nsp15 hexamer forms a platform for engaging dsRNA across multiple protomers. The structures, along with site-directed mutagenesis and RNA cleavage assays revealed critical insight into dsRNA recognition and processing. To process dsRNA Nsp15 utilizes a base-flipping mechanism to properly orient the uridine within the active site for cleavage. Our findings show that Nsp15 is a distinctive endoribonuclease that can cleave both ss- and dsRNA effectively.
Published by Oxford University Press on behalf of Nucleic Acids Research 2022.
Figures


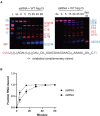


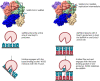
Update of
-
Flipped Over U: Structural Basis for dsRNA Cleavage by the SARS-CoV-2 Endoribonuclease.bioRxiv [Preprint]. 2022 Mar 2:2022.03.02.480688. doi: 10.1101/2022.03.02.480688. bioRxiv. 2022. Update in: Nucleic Acids Res. 2022 Aug 12;50(14):8290-8301. doi: 10.1093/nar/gkac589. PMID: 35262076 Free PMC article. Updated. Preprint.
Similar articles
-
Flipped Over U: Structural Basis for dsRNA Cleavage by the SARS-CoV-2 Endoribonuclease.bioRxiv [Preprint]. 2022 Mar 2:2022.03.02.480688. doi: 10.1101/2022.03.02.480688. bioRxiv. 2022. Update in: Nucleic Acids Res. 2022 Aug 12;50(14):8290-8301. doi: 10.1093/nar/gkac589. PMID: 35262076 Free PMC article. Updated. Preprint.
-
SARS-CoV-2 nsp15 preferentially degrades AU-rich dsRNA via its dsRNA nickase activity.Nucleic Acids Res. 2024 May 22;52(9):5257-5272. doi: 10.1093/nar/gkae290. Nucleic Acids Res. 2024. PMID: 38634805 Free PMC article.
-
Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.Nucleic Acids Res. 2021 Sep 27;49(17):10136-10149. doi: 10.1093/nar/gkab719. Nucleic Acids Res. 2021. PMID: 34403466 Free PMC article.
-
The coronavirus nsp15 endoribonuclease: A puzzling protein and pertinent antiviral drug target.Antiviral Res. 2024 Aug;228:105921. doi: 10.1016/j.antiviral.2024.105921. Epub 2024 May 31. Antiviral Res. 2024. PMID: 38825019 Review.
-
An "Old" protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses.Virology. 2018 Apr;517:157-163. doi: 10.1016/j.virol.2017.12.024. Epub 2018 Jan 4. Virology. 2018. PMID: 29307596 Free PMC article. Review.
Cited by
-
Regulation of coronavirus nsp15 cleavage specificity by RNA structure.PLoS One. 2023 Aug 24;18(8):e0290675. doi: 10.1371/journal.pone.0290675. eCollection 2023. PLoS One. 2023. PMID: 37616296 Free PMC article.
-
SARS-CoV-2 nsp15 enhances viral virulence by subverting host antiviral defenses.Proc Natl Acad Sci U S A. 2025 Jun 17;122(24):e2426528122. doi: 10.1073/pnas.2426528122. Epub 2025 Jun 12. Proc Natl Acad Sci U S A. 2025. PMID: 40504150 Free PMC article.
-
Structural heterogeneity and dynamics in the apical stem loop of s2m from SARS-CoV-2 Delta by an integrative NMR spectroscopy and MD simulation approach.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf552. doi: 10.1093/nar/gkaf552. Nucleic Acids Res. 2025. PMID: 40586311 Free PMC article.
-
Cracking the Code: Enhancing Molecular Tools for Progress in Nanobiotechnology.ACS Appl Bio Mater. 2024 Jun 17;7(6):3587-3604. doi: 10.1021/acsabm.4c00432. Epub 2024 Jun 4. ACS Appl Bio Mater. 2024. PMID: 38833534 Free PMC article. Review.
-
Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.bioRxiv [Preprint]. 2023 Nov 20:2023.11.17.567629. doi: 10.1101/2023.11.17.567629. bioRxiv. 2023. Update in: Protein Cell. 2024 Jul 1;15(7):547-552. doi: 10.1093/procel/pwae009. PMID: 38045375 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

