Restoring and Enhancing the Potency of Existing Antibiotics against Drug-Resistant Gram-Negative Bacteria through the Development of Potent Small-Molecule Adjuvants
- PMID: 35801980
- PMCID: PMC11227883
- DOI: 10.1021/acsinfecdis.2c00121
Restoring and Enhancing the Potency of Existing Antibiotics against Drug-Resistant Gram-Negative Bacteria through the Development of Potent Small-Molecule Adjuvants
Abstract
The rapid and persistent emergence of drug-resistant bacteria poses a looming public health crisis. The possible task of developing new sets of antibiotics to replenish the existing ones is daunting to say the least. Searching for adjuvants that restore or even enhance the potency of existing antibiotics against drug-resistant strains of bacteria represents a practical and cost-effective approach. Herein, we describe the discovery of potent adjuvants that extend the antimicrobial spectrum of existing antibiotics and restore their effectiveness toward drug-resistant strains including mcr-1-expressing strains. From a library of cationic compounds, MD-100, which has a diamidine core structure, was identified as a potent antibiotic adjuvant against Gram-negative bacteria. Further optimization efforts including the synthesis of ∼20 compounds through medicinal chemistry work led to the discovery of a much more potent compound MD-124. MD-124 was shown to sensitize various Gram-negative bacterial species and strains, including multidrug resistant pathogens, toward existing antibiotics with diverse mechanisms of action. We further demonstrated the efficacy of MD-124 in an ex vivo skin infection model and in an in vivo murine systemic infection model using both wild-type and drug-resistant Escherichia coli strains. MD-124 functions through selective permeabilization of the outer membrane of Gram-negative bacteria. Importantly, bacteria exhibited low-resistance frequency toward MD-124. In-depth computational investigations of MD-124 binding to the bacterial outer membrane using equilibrium and steered molecular dynamics simulations revealed key structural features for favorable interactions. The very potent nature of such adjuvants distinguishes them as very useful leads for future drug development in combating bacterial drug resistance.
Keywords: LPS binding; antibiotic adjuvants; effective in vivo; extending the antimicrobial spectrum; overcoming multi-drug resistance.
Conflict of interest statement
Figures



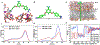




References
-
- Davies SC;Fowler T;Watson J;Livermore DM;Walker D Annual Report of the Chief Medical Officer: infection and the rise of antimicrobial resistance. Lancet 2013, 381 (9878), 1606–1609. - PubMed
-
- Laxminarayan R;Sridhar D;Blaser M;Wang M;Woolhouse M Achieving global targets for antimicrobial resistance. Science 2016, 353 (6302), 874. - PubMed
-
- Blair JMA;Webber MA;Baylay AJ;Ogbolu DO;Piddock LJV Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2014, 13, 42–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

