Type-I-interferon signaling drives microglial dysfunction and senescence in human iPSC models of Down syndrome and Alzheimer's disease
- PMID: 35803230
- PMCID: PMC9345168
- DOI: 10.1016/j.stem.2022.06.007
Type-I-interferon signaling drives microglial dysfunction and senescence in human iPSC models of Down syndrome and Alzheimer's disease
Abstract
Microglia are critical in brain development and Alzheimer's disease (AD) etiology. Down syndrome (DS) is the most common genetic developmental disorder and risk factor for AD. Surprisingly, little information is available on the impact of trisomy of human chromosome 21 (Hsa21) on microglial functions during DS brain development and in AD in DS. Using induced pluripotent stem cell (iPSC)-based organoid and chimeric mouse models, we report that DS microglia exhibit an enhanced synaptic pruning function, which alters neuronal synaptic functions. In response to human brain tissue-derived pathological tau, DS microglia undergo cellular senescence and exhibit elevated type-I-interferon signaling. Mechanistically, knockdown of Hsa21-encoded type I interferon receptors, IFNARs, rescues the DS microglial phenotypes both during brain development and in response to pathological tau. Our findings provide in vivo evidence that human microglia respond to pathological tau by exhibiting dystrophic phenotypes. Targeting IFNARs may improve DS microglial functions and prevent senescence.
Keywords: Alzheimer's disease; Down syndrome; human induced pluripotent stem cells; microglia; senescence; synaptic pruning; type I interferon.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
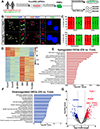
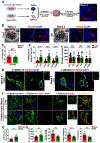
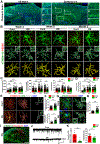
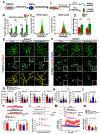


Comment in
-
Human Down syndrome microglia are up for a synaptic feast.Cell Stem Cell. 2022 Jul 7;29(7):1007-1008. doi: 10.1016/j.stem.2022.06.008. Cell Stem Cell. 2022. PMID: 35803219
References
-
- Araya P, Waugh KA, Sullivan KD, Nunez NG, Roselli E, Smith KP, Granrath RE, Rachubinski AL, Enriquez Estrada B, Butcher ET, et al. (2019). Trisomy 21 dysregulates T cell lineages toward an autoimmunity-prone state associated with interferon hyperactivity. Proc Natl Acad Sci U S A 116, 24231–24241. - PMC - PubMed
-
- Bar E, and Barak B (2019). Microglia roles in synaptic plasticity and myelination in homeostatic conditions and neurodevelopmental disorders. Glia 67, 2125–2141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

