Inhibitor induced conformational changes in SARS-COV-2 papain-like protease
- PMID: 35803957
- PMCID: PMC9270405
- DOI: 10.1038/s41598-022-15181-y
Inhibitor induced conformational changes in SARS-COV-2 papain-like protease
Abstract
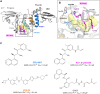
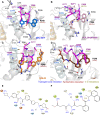
SARS-CoV-2's papain-like protease (PLpro) interaction with ligands has recently been explored with a myriad of crystal structures. We used molecular dynamics (MD) simulations to study different PLpro-ligand complexes, their ligand-induced conformational changes, and interactions. We focused on inhibitors reported with known IC50 against PLpro, namely GRL-0617, XR8-89, PLP_Snyder530, and Sander's recently published compound 7 (CPD7), and compared these trajectories against the apostructure (Apo), with a total of around 60 µs worth simulation data. We aimed to study the conformational changes using molecular dynamics simulations for the inhibitors in the PLpro. PCA analyses and the MSM models revealed distinct conformations of PLpro in the absence/presence of ligands and proposed that BL2-loop contributes to the accessibility of these inhibitors. Further, bulkier substituents closer to Tyr268 and Gln269 could improve inhibition of SARS-CoV-2 PLpro by occupying the region between BL2-groove and BL2-loop, but we also expand on the relevance of exploring multiple PLpro sub-pockets to improve inhibition.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19) - PubMed. https://pubmed.ncbi.nlm.nih.gov/32238757/. - PMC - PubMed
-
- An overview of the epidemiologic, diagnostic and treatment approaches of COVID-19: What do we know? https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8245675/. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

