J-SPACE: a Julia package for the simulation of spatial models of cancer evolution and of sequencing experiments
- PMID: 35804300
- PMCID: PMC9270769
- DOI: 10.1186/s12859-022-04779-8
J-SPACE: a Julia package for the simulation of spatial models of cancer evolution and of sequencing experiments
Abstract
Background: The combined effects of biological variability and measurement-related errors on cancer sequencing data remain largely unexplored. However, the spatio-temporal simulation of multi-cellular systems provides a powerful instrument to address this issue. In particular, efficient algorithmic frameworks are needed to overcome the harsh trade-off between scalability and expressivity, so to allow one to simulate both realistic cancer evolution scenarios and the related sequencing experiments, which can then be used to benchmark downstream bioinformatics methods.
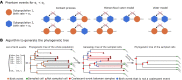
Result: We introduce a Julia package for SPAtial Cancer Evolution (J-SPACE), which allows one to model and simulate a broad set of experimental scenarios, phenomenological rules and sequencing settings.Specifically, J-SPACE simulates the spatial dynamics of cells as a continuous-time multi-type birth-death stochastic process on a arbitrary graph, employing different rules of interaction and an optimised Gillespie algorithm. The evolutionary dynamics of genomic alterations (single-nucleotide variants and indels) is simulated either under the Infinite Sites Assumption or several different substitution models, including one based on mutational signatures. After mimicking the spatial sampling of tumour cells, J-SPACE returns the related phylogenetic model, and allows one to generate synthetic reads from several Next-Generation Sequencing (NGS) platforms, via the ART read simulator. The results are finally returned in standard FASTA, FASTQ, SAM, ALN and Newick file formats.
Conclusion: J-SPACE is designed to efficiently simulate the heterogeneous behaviour of a large number of cancer cells and produces a rich set of outputs. Our framework is useful to investigate the emergent spatial dynamics of cancer subpopulations, as well as to assess the impact of incomplete sampling and of experiment-specific errors. Importantly, the output of J-SPACE is designed to allow the performance assessment of downstream bioinformatics pipelines processing NGS data. J-SPACE is freely available at: https://github.com/BIMIB-DISCo/J-Space.jl .
Keywords: Cancer Evolution; Next-generation sequencing; Spatial dynamics; Stochastic Simulation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

