getSequenceInfo: a suite of tools allowing to get genome sequence information from public repositories
- PMID: 35804320
- PMCID: PMC9264741
- DOI: 10.1186/s12859-022-04809-5
getSequenceInfo: a suite of tools allowing to get genome sequence information from public repositories
Abstract
Background: Biological sequences are increasing rapidly and exponentially worldwide. Nucleotide sequence databases play an important role in providing meaningful genomic information on a variety of biological organisms.
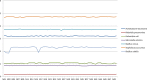
Results: The getSequenceInfo software tool allows to access sequence information from various public repositories (GenBank, RefSeq, and the European Nucleotide Archive), and is compatible with different operating systems (Linux, MacOS, and Microsoft Windows) in a programmatic way (command line) or as a graphical user interface. getSequenceInfo or gSeqI v1.0 should help users to get some information on queried sequences that could be useful for specific studies (e.g. the country of origin/isolation or the release date of queried sequences). Queries can be made to retrieve sequence data based on a given kingdom and species, or from a given date. This program allows the separation between chromosomes and plasmids (or other genetic elements/components) by arranging each component in a given folder. Some basic statistics are also performed by the program (such as the calculation of GC content for queried assemblies). An empirically designed nucleotide ratio is calculated using nucleotide information in order to tentatively provide a "NucleScore" for studied genome assemblies. Besides the main gSeqI tool, other additional tools have been developed to perform various tasks related to sequence analysis.
Conclusion: The aim of this study is to democratize the use of public repositories in programmatic ways, and to facilitate sequence data analysis in a pedagogical perspective. Output results are available in FASTA, FASTQ, Excel/TSV or HTML formats. The program is freely available at: https://github.com/karubiotools/getSequenceInfo . getSequenceInfo and supplementary tools are partly available through the recently released Galaxy KaruBioNet platform ( http://calamar.univ-ag.fr/c3i/galaxy_karubionet.html ).
Keywords: Assembly; DNA; Genome sequences; Metadata; Nucleotide diversity; Repository.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Amid C, Alako BTF, BalavenkataramanKadhirvelu V, Burdett T, Burgin J, Fan J, Harrison PW, Holt S, Hussein A, Ivanov E, Jayathilaka S, Kay S, Keane T, Leinonen R, Liu X, Martinez-Villacorta J, Milano A, Pakseresht A, Rahman N, Rajan J, Reddy K, Richards E, Smirnov D, Sokolov A, Vijayaraja S, Cochrane G. The European nucleotide archive in 2019. Nucleic Acids Res. 2020;48:D70–D76. doi: 10.1093/nar/gkz1063. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

