Pre-PCR Mutation-Enrichment Methods for Liquid Biopsy Applications
- PMID: 35804916
- PMCID: PMC9264780
- DOI: 10.3390/cancers14133143
Pre-PCR Mutation-Enrichment Methods for Liquid Biopsy Applications
Abstract
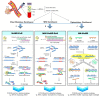
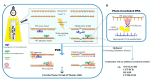
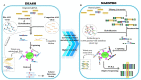
Liquid biopsy is having a remarkable impact on healthcare- and disease-management in the context of personalized medicine. Circulating free DNA (cfDNA) is one of the most instructive liquid-biopsy-based biomarkers and harbors valuable information for diagnostic, predictive, and prognostic purposes. When it comes to cancer, circulating DNA from the tumor (ctDNA) has a wide range of applications, from early cancer detection to the early detection of relapse or drug resistance, and the tracking of the dynamic genomic make-up of tumor cells. However, the detection of ctDNA remains technically challenging, due, in part, to the low frequency of ctDNA among excessive circulating cfDNA originating from normal tissues. During the past three decades, mutation-enrichment methods have emerged to boost sensitivity and enable facile detection of low-level mutations. Although most developed techniques apply mutation enrichment during or following initial PCR, there are a few techniques that allow mutation selection prior to PCR, which provides advantages. Pre-PCR enrichment techniques can be directly applied to genomic DNA and diminish the influence of PCR errors that can take place during amplification. Moreover, they have the capability for high multiplexity and can be followed by established mutation detection and enrichment technologies without changes to their established procedures. The first approaches for pre-PCR enrichment were developed by employing restriction endonucleases directly on genomic DNA in the early 1990s. However, newly developed pre-PCR enrichment methods provide higher sensitivity and versatility. This review describes the available pre-PCR enrichment methods and focuses on the most recently developed techniques (NaME-PrO, UVME, and DEASH/MAESTRO), emphasizing their applications in liquid biopsies.
Keywords: cell-free DNA; circulating tumor DNA; liquid biopsy; low-level mutation detection; mutation enrichment; pre-PCR enrichment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Recent Developments in Mutation Enrichment and Detection Technologies.Clin Chem. 2022 Oct 6;68(10):1250-1260. doi: 10.1093/clinchem/hvac093. Clin Chem. 2022. PMID: 35716101 Review.
-
Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management.Comput Struct Biotechnol J. 2018 Oct 9;16:370-378. doi: 10.1016/j.csbj.2018.10.002. eCollection 2018. Comput Struct Biotechnol J. 2018. PMID: 30364656 Free PMC article. Review.
-
Multiplex detection of ctDNA mutations in plasma of colorectal cancer patients by PCR/SERS assay.Nanotheranostics. 2020 Aug 25;4(4):224-232. doi: 10.7150/ntno.48905. eCollection 2020. Nanotheranostics. 2020. PMID: 32923312 Free PMC article.
-
A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications.Genes Chromosomes Cancer. 2018 Mar;57(3):123-139. doi: 10.1002/gcc.22517. Epub 2017 Dec 20. Genes Chromosomes Cancer. 2018. PMID: 29205637 Review.
-
Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies.Lab Chip. 2018 Apr 17;18(8):1174-1196. doi: 10.1039/C8LC00100F. Lab Chip. 2018. PMID: 29569666 Review.
Cited by
-
Development and validation of a multi-marker liquid bead array assay for the simultaneous detection of PIK3CA and ESR1 hotspot mutations in single circulating tumor cells (CTCs).Heliyon. 2024 Sep 12;10(19):e37873. doi: 10.1016/j.heliyon.2024.e37873. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39386783 Free PMC article.
-
Extrinsic and intrinsic preanalytical variables affecting liquid biopsy in cancer.Cell Rep Med. 2023 Oct 17;4(10):101196. doi: 10.1016/j.xcrm.2023.101196. Epub 2023 Sep 18. Cell Rep Med. 2023. PMID: 37725979 Free PMC article. Review.
-
A panorama of colon cancer in the era of liquid biopsy.J Liq Biopsy. 2024 Mar 13;4:100148. doi: 10.1016/j.jlb.2024.100148. eCollection 2024 Jun. J Liq Biopsy. 2024. PMID: 40027146 Free PMC article. Review.
-
Clinical Utility and Application of Liquid Biopsy Genotyping in Lung Cancer: A Comprehensive Review.Lung Cancer (Auckl). 2023 Feb 3;14:11-25. doi: 10.2147/LCTT.S388047. eCollection 2023. Lung Cancer (Auckl). 2023. PMID: 36762267 Free PMC article. Review.
-
Enzymatic Methods for Mutation Detection in Cancer Samples and Liquid Biopsies.Int J Mol Sci. 2023 Jan 4;24(2):923. doi: 10.3390/ijms24020923. Int J Mol Sci. 2023. PMID: 36674433 Free PMC article. Review.
References
-
- Giusti I., Bianchi S., Nottola S.A., Macchiarelli G., Dolo V. Clinical Electron Microscopy in the Study of Human Ovarian Tissues. EuroMediterr. Biomed. J. 2019;14:145–151.
-
- Alix-Panabieres C., Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

