Multiplex Tissue Imaging: Spatial Revelations in the Tumor Microenvironment
- PMID: 35804939
- PMCID: PMC9264815
- DOI: 10.3390/cancers14133170
Multiplex Tissue Imaging: Spatial Revelations in the Tumor Microenvironment
Abstract
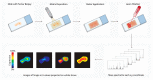
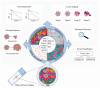
The tumor microenvironment is a complex ecosystem containing various cell types, such as immune cells, fibroblasts, and endothelial cells, which interact with the tumor cells. In recent decades, the cancer research field has gained insight into the cellular subtypes that are involved in tumor microenvironment heterogeneity. Moreover, it has become evident that cellular interactions in the tumor microenvironment can either promote or inhibit tumor development, progression, and drug resistance, depending on the context. Multiplex spatial analysis methods have recently been developed; these have offered insight into how cellular crosstalk dynamics and heterogeneity affect cancer prognoses and responses to treatment. Multiplex (imaging) technologies and computational analysis methods allow for the spatial visualization and quantification of cell-cell interactions and properties. These technological advances allow for the discovery of cellular interactions within the tumor microenvironment and provide detailed single-cell information on properties that define cellular behavior. Such analyses give insights into the prognosis and mechanisms of therapy resistance, which is still an urgent problem in the treatment of multiple types of cancer. Here, we provide an overview of multiplex imaging technologies and concepts of downstream analysis methods to investigate cell-cell interactions, how these studies have advanced cancer research, and their potential clinical implications.
Keywords: MALDI-MSI; MIBI; PhenoCycler; PhenoImager; cancer; imaging mass cytometry; multiplex imaging; single-cell data analysis; spatial analysis; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Elaldi R., Hemon P., Petti L., Cosson E., Desrues B., Sudaka A., Poissonnet G., Van Obberghen-Schilling E., Pers J.-O., Braud V.M., et al. High Dimensional Imaging Mass Cytometry Panel to Visualize the Tumor Immune Microenvironment Contexture. Front. Immunol. 2021;12:666233. doi: 10.3389/fimmu.2021.666233. - DOI - PMC - PubMed
-
- Li H., van der Leun A.M., Yofe I., Lubling Y., Gelbard-Solodkin D., van Akkooi A.C., van den Braber M., Rozeman E.A., Haanen J.B., Blank C.U., et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 2020;181:747. doi: 10.1016/j.cell.2020.04.017. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

