Novel Approaches in Molecular Characterization of Classical Hodgkin Lymphoma
- PMID: 35805000
- PMCID: PMC9264882
- DOI: 10.3390/cancers14133222
Novel Approaches in Molecular Characterization of Classical Hodgkin Lymphoma
Abstract
Classical Hodgkin lymphoma (cHL) represents a B-cell lymphoproliferative disease characterized by clonal immunoglobulin gene rearrangements and recurrent genomic aberrations in the Hodgkin Reed-Sternberg cells in a reactive inflammatory background. Several methods are available for the molecular analysis of cHL on both tissue and cell-free DNA isolated from blood, which can provide detailed information regarding the clonal composition and genetic alterations that drive lymphoma pathogenesis. Clonality testing involving the detection of immunoglobulin and T cell receptor gene rearrangements, together with mutation analysis, represent valuable tools for cHL diagnostics, especially for patients with an atypical histological or clinical presentation reminiscent of a reactive lesion or another lymphoma subtype. In addition, clonality assessment may establish the clonal relationship of composite or subsequent lymphoma presentations within one patient. During the last few decades, more insight has been obtained on the molecular mechanisms that drive cHL development, including recurrently affected signaling pathways (e.g., NF-κB and JAK/STAT) and immune evasion. We provide an overview of the different approaches to characterize the molecular composition of cHL, and the implementation of these next-generation sequencing-based techniques in research and diagnostic settings.
Keywords: cell-free DNA; classical Hodgkin lymphoma; clonality assessment; mutation analysis; next-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
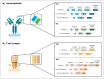

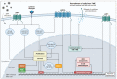
References
-
- Swerdlow S.H., Campo E., Harris N.L., Pileri S.A. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. International Agency for Research on Cancer; Lyon, France: 2017. Myeloproliferative neoplasms.
-
- Campo E., Harris N.L., Pileri S.A., Jaffe E.S., Stein H., Thiele J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. International Agency for Research on Cancer; Lyon, France: 2017. Mature B-cell neoplasms.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

