iPCD: A Comprehensive Data Resource of Regulatory Proteins in Programmed Cell Death
- PMID: 35805101
- PMCID: PMC9265749
- DOI: 10.3390/cells11132018
iPCD: A Comprehensive Data Resource of Regulatory Proteins in Programmed Cell Death
Abstract
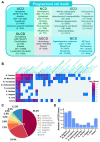
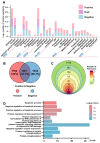
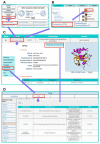
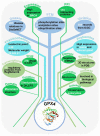
Programmed cell death (PCD) is an essential biological process involved in many human pathologies. According to the continuous discovery of new PCD forms, a large number of proteins have been found to regulate PCD. Notably, post-translational modifications play critical roles in PCD process and the rapid advances in proteomics have facilitated the discovery of new PCD proteins. However, an integrative resource has yet to be established for maintaining these regulatory proteins. Here, we briefly summarize the mainstream PCD forms, as well as the current progress in the development of public databases to collect, curate and annotate PCD proteins. Further, we developed a comprehensive database, with integrated annotations for programmed cell death (iPCD), which contained 1,091,014 regulatory proteins involved in 30 PCD forms across 562 eukaryotic species. From the scientific literature, we manually collected 6493 experimentally identified PCD proteins, and an orthologous search was then conducted to computationally identify more potential PCD proteins. Additionally, we provided an in-depth annotation of PCD proteins in eight model organisms, by integrating the knowledge from 102 additional resources that covered 16 aspects, including post-translational modification, protein expression/proteomics, genetic variation and mutation, functional annotation, structural annotation, physicochemical property, functional domain, disease-associated information, protein-protein interaction, drug-target relation, orthologous information, biological pathway, transcriptional regulator, mRNA expression, subcellular localization and DNA and RNA element. With a data volume of 125 GB, we anticipate that iPCD can serve as a highly useful resource for further analysis of PCD in eukaryotes.
Keywords: GPX4; PCD protein; apoptosis; ferroptosis; programmed cell death.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

