Insights into the Structure and Function of the Pex1/Pex6 AAA-ATPase in Peroxisome Homeostasis
- PMID: 35805150
- PMCID: PMC9265785
- DOI: 10.3390/cells11132067
Insights into the Structure and Function of the Pex1/Pex6 AAA-ATPase in Peroxisome Homeostasis
Abstract
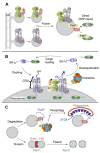
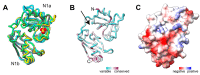
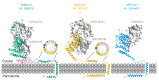
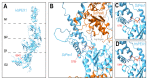
The AAA-ATPases Pex1 and Pex6 are required for the formation and maintenance of peroxisomes, membrane-bound organelles that harbor enzymes for specialized metabolism. Together, Pex1 and Pex6 form a heterohexameric AAA-ATPase capable of unfolding substrate proteins via processive threading through a central pore. Here, we review the proposed roles for Pex1/Pex6 in peroxisome biogenesis and degradation, discussing how the unfolding of potential substrates contributes to peroxisome homeostasis. We also consider how advances in cryo-EM, computational structure prediction, and mechanisms of related ATPases are improving our understanding of how Pex1/Pex6 converts ATP hydrolysis into mechanical force. Since mutations in PEX1 and PEX6 cause the majority of known cases of peroxisome biogenesis disorders such as Zellweger syndrome, insights into Pex1/Pex6 structure and function are important for understanding peroxisomes in human health and disease.
Keywords: AAA-ATPase; PEX1; PEX26; PEX6; organelle biogenesis; peroxisomes; translocation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
The N1 domain of the peroxisomal AAA-ATPase Pex6 is required for Pex15 binding and proper assembly with Pex1.J Biol Chem. 2024 Jan;300(1):105504. doi: 10.1016/j.jbc.2023.105504. Epub 2023 Nov 29. J Biol Chem. 2024. PMID: 38036174 Free PMC article.
-
The peroxisomal AAA-ATPase Pex1/Pex6 unfolds substrates by processive threading.Nat Commun. 2018 Jan 10;9(1):135. doi: 10.1038/s41467-017-02474-4. Nat Commun. 2018. PMID: 29321502 Free PMC article.
-
The Pex1/Pex6 complex is a heterohexameric AAA+ motor with alternating and highly coordinated subunits.J Mol Biol. 2015 Mar 27;427(6 Pt B):1375-1388. doi: 10.1016/j.jmb.2015.01.019. Epub 2015 Feb 7. J Mol Biol. 2015. PMID: 25659908 Free PMC article.
-
Structures of the double-ring AAA ATPase Pex1-Pex6 involved in peroxisome biogenesis.FEBS J. 2016 Mar;283(6):986-92. doi: 10.1111/febs.13569. Epub 2015 Nov 12. FEBS J. 2016. PMID: 26476099 Free PMC article. Review.
-
Structural Mapping of Missense Mutations in the Pex1/Pex6 Complex.Int J Mol Sci. 2019 Aug 1;20(15):3756. doi: 10.3390/ijms20153756. Int J Mol Sci. 2019. PMID: 31374812 Free PMC article. Review.
Cited by
-
Towards solving the mystery of peroxisomal matrix protein import.Trends Cell Biol. 2024 May;34(5):388-405. doi: 10.1016/j.tcb.2023.08.005. Epub 2023 Sep 22. Trends Cell Biol. 2024. PMID: 37743160 Free PMC article. Review.
-
Distinct and Shared Molecular Mechanisms in Pediatric Antrochoanal Polyps and Chronic Rhinosinusitis with Nasal Polyps: A Proteomic and Metabolomic Integrative Analysis.J Inflamm Res. 2025 Mar 26;18:4435-4447. doi: 10.2147/JIR.S507475. eCollection 2025. J Inflamm Res. 2025. PMID: 40166594 Free PMC article.
-
A century of anthropogenic perturbations impact genomic signatures of the iconic migratory Atlantic cod.Sci Adv. 2025 Aug;11(31):eadp3342. doi: 10.1126/sciadv.adp3342. Epub 2025 Jul 30. Sci Adv. 2025. PMID: 40737403 Free PMC article.
-
Identification of Potential Feature Genes in CRSwNP Using Bioinformatics Analysis and Machine Learning Strategies.J Inflamm Res. 2024 Oct 22;17:7573-7590. doi: 10.2147/JIR.S484914. eCollection 2024. J Inflamm Res. 2024. PMID: 39464338 Free PMC article.
-
Arrayed CRISPR libraries for the genome-wide activation, deletion and silencing of human protein-coding genes.Nat Biomed Eng. 2025 Jan;9(1):127-148. doi: 10.1038/s41551-024-01278-4. Epub 2024 Dec 4. Nat Biomed Eng. 2025. PMID: 39633028 Free PMC article.

