Lmx1a-Dependent Activation of miR-204/211 Controls the Timing of Nurr1-Mediated Dopaminergic Differentiation
- PMID: 35805964
- PMCID: PMC9266978
- DOI: 10.3390/ijms23136961
Lmx1a-Dependent Activation of miR-204/211 Controls the Timing of Nurr1-Mediated Dopaminergic Differentiation
Abstract
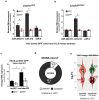
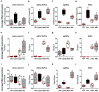
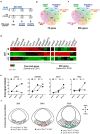
The development of midbrain dopaminergic (DA) neurons requires a fine temporal and spatial regulation of a very specific gene expression program. Here, we report that during mouse brain development, the microRNA (miR-) 204/211 is present at a high level in a subset of DA precursors expressing the transcription factor Lmx1a, an early determinant for DA-commitment, but not in more mature neurons expressing Th or Pitx3. By combining different in vitro model systems of DA differentiation, we show that the levels of Lmx1a influence the expression of miR-204/211. Using published transcriptomic data, we found a significant enrichment of miR-204/211 target genes in midbrain dopaminergic neurons where Lmx1a was selectively deleted at embryonic stages. We further demonstrated that miR-204/211 controls the timing of the DA differentiation by directly downregulating the expression of Nurr1, a late DA differentiation master gene. Thus, our data indicate the Lmx1a-miR-204/211-Nurr1 axis as a key component in the cascade of events that ultimately lead to mature midbrain dopaminergic neurons differentiation and point to miR-204/211 as the molecular switch regulating the timing of Nurr1 expression.
Keywords: Lmx1a; Nurr1; dopamine; microRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Volpicelli F., Perrone-Capano C., Bellenchi G.C., Colucci-D’Amato L., di Porzio U. Molecular Regulation in Dopaminergic Neuron Development. Cues to Unveil Molecular Pathogenesis and Pharmacological Targets of Neurodegeneration. Int. J. Mol. Sci. 2020;21:3995. doi: 10.3390/ijms21113995. - DOI - PMC - PubMed
-
- Lo Iacono L., Catale C., Martini A., Valzania A., Viscomi M.T., Chiurchiù V., Guatteo E., Bussone S., Perrone F., Di Sabato P., et al. From Traumatic Childhood to Cocaine Abuse: The Critical Function of the Immune System. Biol. Psychiatry. 2018;84:905–916. doi: 10.1016/j.biopsych.2018.05.022. - DOI - PubMed
-
- D’Addario S.L., Di Segni M., Ledonne A., Piscitelli R., Babicola L., Martini A., Spoleti E., Mancini C., Ielpo D., D’Amato F.R., et al. Resilience to Anhedonia-Passive Coping Induced by Early Life Experience Is Linked to a Long-Lasting Reduction of Ih Current in VTA Dopaminergic Neurons. Neurobiol. Stress. 2021;14:100324. doi: 10.1016/j.ynstr.2021.100324. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

