Computational Analysis Identifies Novel Biomarkers for High-Risk Bladder Cancer Patients
- PMID: 35806060
- PMCID: PMC9266725
- DOI: 10.3390/ijms23137057
Computational Analysis Identifies Novel Biomarkers for High-Risk Bladder Cancer Patients
Abstract
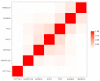
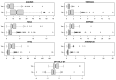
In the case of bladder cancer, carcinoma in situ (CIS) is known to have poor diagnosis. However, there are not enough studies that examine the biomarkers relevant to CIS development. Omics experiments generate data with tens of thousands of descriptive variables, e.g., gene expression levels. Often, many of these descriptive variables are identified as somehow relevant, resulting in hundreds or thousands of relevant variables for building models or for further data analysis. We analyze one such dataset describing patients with bladder cancer, mostly non-muscle-invasive (NMIBC), and propose a novel approach to feature selection. This approach returns high-quality features for prediction and yet allows interpretability as well as a certain level of insight into the analyzed data. As a result, we obtain a small set of seven of the most-useful biomarkers for diagnostics. They can also be used to build tests that avoid the costly and time-consuming existing methods. We summarize the current biological knowledge of the chosen biomarkers and contrast it with our findings.
Keywords: biomarker identification; carcinoma in situ (CIS); nonmuscle-invasive bladder cancer (NMIBC); optimal feature set selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
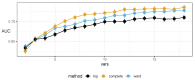

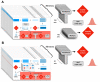
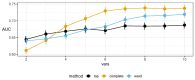

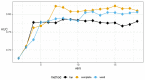
References
-
- Chen J., Zhang H., Sun G., Zhang X., Zhao J., Liu J., Shen P., Shi M., Zeng H. Comparison of the prognosis of primary and progressive muscle-invasive bladder cancer after radical cystectomy: A systematic review and meta-analysis. Int. J. Surg. 2018;52:214–220. doi: 10.1016/j.ijsu.2018.02.049. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

