MGMT and Whole-Genome DNA Methylation Impacts on Diagnosis, Prognosis and Therapy of Glioblastoma Multiforme
- PMID: 35806153
- PMCID: PMC9266959
- DOI: 10.3390/ijms23137148
MGMT and Whole-Genome DNA Methylation Impacts on Diagnosis, Prognosis and Therapy of Glioblastoma Multiforme
Abstract
Epigenetic changes in DNA methylation contribute to the development of many diseases, including cancer. In glioblastoma multiforme, the most prevalent primary brain cancer and an incurable tumor with a median survival time of 15 months, a single epigenetic modification, the methylation of the O6-Methylguanine-DNA Methyltransferase (MGMT) gene, is a valid biomarker for predicting response to therapy with alkylating agents and also, independently, prognosis. More recently, the progress from single gene to whole-genome analysis of DNA methylation has allowed a better subclassification of glioblastomas. Here, we review the clinically relevant information that can be obtained by studying MGMT gene and whole-genome DNA methylation changes in glioblastomas, also highlighting benefits, including those of liquid biopsy, and pitfalls of the different detection methods. Finally, we discuss how changes in DNA methylation, especially in glioblastomas bearing mutations in the Isocitrate Dehydrogenase (IDH) 1 and 2 genes, can be exploited as targets for tailoring therapy.
Keywords: IDH1/2 mutations; MGMT gene methylation; glioblastoma multiforme; liquid biopsy; whole-genome methylation profiling.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript.
Figures

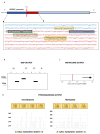
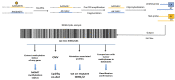
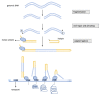
References
-
- Friedman H.S., Kerby T., Calvert H. Temozolomide and treatment of malignant glioma. Clin. Cancer Res. 2000;6:2585–2597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

