Transcriptomic and Metabolomic Analysis of Quality Changes during Sweet Cherry Fruit Development and Mining of Related Genes
- PMID: 35806406
- PMCID: PMC9266358
- DOI: 10.3390/ijms23137402
Transcriptomic and Metabolomic Analysis of Quality Changes during Sweet Cherry Fruit Development and Mining of Related Genes
Abstract
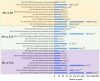
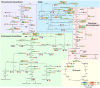
Sweet cherries are economically important fruit trees, and their quality changes during development need to be determined. The mechanism of fruit quality changes in sweet cherries were determined by analyzing sweet cherry fruits at 12 developmental stages. The results showed that the soluble sugar, anthocyanin content, and hormones of sweet cherries all changed drastically during the color transition. Therefore, the fruits at the beginning of color conversion, at the end of color conversion, and at the ripening state were selected for the comprehensive analysis of their metabolome and transcriptome. Different sugars, such as D-glucose, sucrose, and trehalose, were identified in the metabolome. Dihydroquercetin, delphinidin-3-glucoside, cyanidin-3-rutincoside, and other flavonoid species were also identified. D-glucose and cyanidin-3-rutinoside were among the most important components of sweet cherry soluble sugars and anthocyanins, respectively. The transcriptional analysis identified key structural genes and nine transcription factors involved in the ABA, sugar, organic acid, and anthocyanin synthesis pathways, with the following specific regulatory patterns. NAC71, WRKY57, and WRKY3 regulate fruit sugar accumulation mainly by acting on INV, SPS, and SUS. MYC2 is involved in the synthesis of anthocyanin precursors by activating PAL and C4H, whereas TCP7 mainly regulates CHI and F3H. WRKY3, NAC71, and WRKY57 have important positive regulatory significance on anthocyanin accumulation, mainly by activating the expression of DFR, ANS, and 3GT.
Keywords: fruit development; metabolome; quality formation; sweet cherry; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Valero D., Serrano M. Growth and ripening stage at harvest modulates postharvest quality and bioactive compounds with antioxidant activity. Stewart Postharvest Rev. 2013;3:1–8. doi: 10.2212/spr.2013.3.7. - DOI
-
- Kader A.A. Fruit maturity, ripening, and quality relationships. International Symposium Effect of Pre-& Postharvest Factors in Fruit Storage. Acta Hortic. 1999;485:203–208. doi: 10.17660/ActaHortic.1999.485.27. - DOI
-
- Alonso M.A.S., Paquin J.P., Mangin J.P.L. Modelling perceived quality in fruit products: Their extrinsic and intrinsic attributes. J. Food Prod Mark. 2002;8:29–48. doi: 10.1300/J038v08n01_03. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

