Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
- PMID: 35809383
- PMCID: PMC9259064
- DOI: 10.1016/j.jmb.2022.167706
Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
Abstract
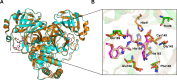
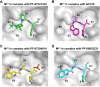
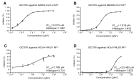
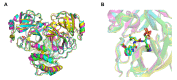
New variants of the severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) emerged and spread rapidly all over the world, which strongly supports the need for pharmacological options to complement vaccine strategies. Main protease (Mpro or 3CLpro) is a critical enzyme in the life cycle of SARS-CoV-2 and appears to be highly conserved among different genera of coronaviruses, making it an ideal target for the development of drugs with broad-spectrum property. PF-07304814 developed by Pfizer is an intravenously administered inhibitor targeting SARS-CoV-2 Mpro. Here we showed that PF-07304814 displays broad-spectrum inhibitory activity against Mpros from multiple coronaviruses. Crystal structures of Mpros of SARS-CoV-2, SARS-CoV, MERS-CoV, and HCoV-NL63 bound to the inhibitor PF-07304814 revealed a conserved ligand-binding site, providing new insights into the mechanism of inhibition of viral replication. A detailed analysis of these crystal structures complemented by comprehensive comparison defined the key structural determinants essential for inhibition and illustrated the binding mode of action of Mpros from different coronaviruses. In view of the importance of Mpro for the medications of SARS-CoV-2 infection, insights derived from the present study should accelerate the design of pan-coronaviral main protease inhibitors that are safer and more effective.
Keywords: PF-07304814; coronavirus; inhibitor; main protease; structural basis.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no conflict of interest.
Figures


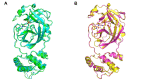

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

