Significant Subgraph Detection in Multi-omics Networks for Disease Pathway Identification
- PMID: 35811829
- PMCID: PMC9256965
- DOI: 10.3389/fdata.2022.894632
Significant Subgraph Detection in Multi-omics Networks for Disease Pathway Identification
Abstract
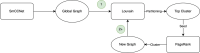
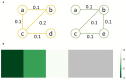
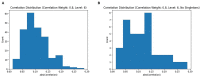
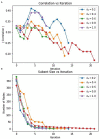
Chronic obstructive pulmonary disease (COPD) is one of the leading causes of death in the United States. COPD represents one of many areas of research where identifying complex pathways and networks of interacting biomarkers is an important avenue toward studying disease progression and potentially discovering cures. Recently, sparse multiple canonical correlation network analysis (SmCCNet) was developed to identify complex relationships between omics associated with a disease phenotype, such as lung function. SmCCNet uses two sets of omics datasets and an associated output phenotypes to generate a multi-omics graph, which can then be used to explore relationships between omics in the context of a disease. Detecting significant subgraphs within this multi-omics network, i.e., subgraphs which exhibit high correlation to a disease phenotype and high inter-connectivity, can help clinicians identify complex biological relationships involved in disease progression. The current approach to identifying significant subgraphs relies on hierarchical clustering, which can be used to inform clinicians about important pathways involved in the disease or phenotype of interest. The reliance on a hierarchical clustering approach can hinder subgraph quality by biasing toward finding more compact subgraphs and removing larger significant subgraphs. This study aims to introduce new significant subgraph detection techniques. In particular, we introduce two subgraph detection methods, dubbed Correlated PageRank and Correlated Louvain, by extending the Personalized PageRank Clustering and Louvain algorithms, as well as a hybrid approach combining the two proposed methods, and compare them to the hierarchical method currently in use. The proposed methods show significant improvement in the quality of the subgraphs produced when compared to the current state of the art.
Keywords: Louvain; PageRank; graph clustering; multi-omics graph; subgraph detection.
Copyright © 2022 Abdel-Hafiz, Najafi, Helmi, Pratte, Zhuang, Liu, Kechris, Bowler, Lange and Banaei-Kashani.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Baadel S., Thabtah F., Lu J. (2016). Overlapping clustering: a review. 2016 SAI Computing Conference (SAI). London. 10.1109/SAI.2016.7555988 - DOI
-
- Bhatt S., Padhee S., Sheth A., Chen K., Shalin V., Doran D., et al. . (2019). Knowledge Graph Enhanced Community Detection and Characterization. New York, NY: Association for Computing Machinery. 10.1145/3289600.3291031 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

