Genome-wide Meta-analysis Reveals New Gene Signatures and Potential Drug Targets of Hypertension
- PMID: 35811894
- PMCID: PMC9260904
- DOI: 10.1021/acsomega.2c02277
Genome-wide Meta-analysis Reveals New Gene Signatures and Potential Drug Targets of Hypertension
Abstract
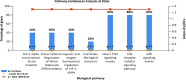
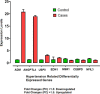
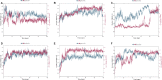
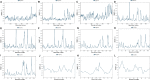
The prevalence of hypertension reported around the world is increasing and is an important public health challenge. This study was designed to explore the disease's genetic variations and to identify new hypertension-related genes and target proteins. We analyzed 22 publicly available Affymetrix cDNA datasets of hypertension using an integrated system-level framework involving differential expression genetic (DEG) analysis, data mining, gene enrichment, protein-protein interaction, microRNA analysis, toxicogenomics, gene regulation, molecular docking, and simulation studies. We found potential DEGs after screening out the extracellular proteins. We studied the functional role of seven shortlisted DEGs (ADM, EDN1, ANGPTL4, NFIL3, MSR1, CEBPD, and USP8) in hypertension after disease gene curation analysis. The expression profiling and cluster analysis showed significant variations and enriched GO terms. hsa-miR-365a-3p, hsa-miR-2052, hsa-miR-3065-3p, hsa-miR-603, hsa-miR-7113-3p, hsa-miR-3923, and hsa-miR-524-5p were identified as hypertension-associated miRNA targets for each gene using computational algorithms. We found functional interactions of source DEGs with target and important gene signatures including EGFR, AGT, AVP, APOE, RHOA, SRC, APOB, STAT3, UBC, LPL, APOA1, and AKT1 associated with the disease. These DEGs are mainly involved in fatty acid metabolism, myometrial pathways, MAPK, and G-alpha signaling pathways linked with hypertension pathogenesis. We predicted significantly disordered regions of 71.2, 48.8, and 45.4% representing the mutation in the sequence of NFIL3, USP8, and ADM, respectively. Regulation of gene expression was performed to find upregulated genes. Molecular docking analysis was used to evaluate Food and Drug Administration-approved medicines against the four DEGs that were overexpressed. For each elevated target protein, the three best drug candidates were chosen. Furthermore, molecular dynamics (MD) simulation using the target's active sites for 100 ns was used to validate these 12 complexes after docking. This investigation establishes the worth of systems genetics for finding four possible genes as potential drug targets for hypertension. These network-based approaches are significant for finding genetic variant data, which will advance the understanding of how to hasten the identification of drug targets and improve the understanding regarding the treatment of hypertension.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures














References
-
- Singh R.; Suh I.; Singh V.; Chaithiraphan S.; Laothavorn P.; Sy R.; Babilonia N.; Rahman A.; Sheikh S.; Tomlinson B.; Sarraf-Zadigan N. Hypertension and stroke in Asia: prevalence, control and strategies in developing countries for prevention. J. Hum. Hypertens. 2000, 14, 749–763. 10.1038/sj.jhh.1001057. - DOI - PubMed
-
- Gupta R. Hypertension in India--definition, prevalence and evaluation. J. Indian Med. Assoc. 1999, 97, 74–80. - PubMed
-
- Mills K. T.; Bundy J. D.; Kelly T. N.; Reed J. E.; Kearney P. M.; Reynolds K.; Chen J.; He J. Global disparities of hypertension prevalence and control: a systematic analysis of population-based studies from 90 countries. Circulation 2016, 134, 441–450. 10.1161/CIRCULATIONAHA.115.018912. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

