DDOST Correlated with Malignancies and Immune Microenvironment in Gliomas
- PMID: 35812432
- PMCID: PMC9260604
- DOI: 10.3389/fimmu.2022.917014
DDOST Correlated with Malignancies and Immune Microenvironment in Gliomas
Abstract
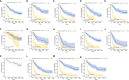
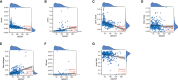
Among the most common types of brain tumor, gliomas are the most aggressive and have the poorest prognosis. Dolichyl-diphosphooligosaccharide protein glycosyltransferase non-catalytic subunit (DDOST) encodes a component of the oligosaccharide transferase complex and is related to the N-glycosylation of proteins. The role of DDOST in gliomas, however, is not yet known. First, we performed a pan cancer analysis of DDOST in the TCGA cohort. The expression of DDOST was compared between glioma and normal brain tissues in the GEO and Chinese Glioma Genome Atlas (CGGA) databases. In order to explore the role of DDOST in glioma, we analyze the impact of DDOST on the prognosis of glioma patients, with the CGGA 325 dataset as a test set and the CGGA 693 dataset as a validation set. Immunohistochemistry was performed on tissue microarrays to examine whether DDOST has an impact on glioma patient survival. Next, using single-cell sequencing analysis, GSEA, immune infiltration analysis, and mutation analysis, we explored how DDOST affected the glioma tumor microenvironment. Finally, we evaluated the clinical significance of DDOST for glioma treatment by constructing nomograms and decision curve analysis (DCA) curves. We found that DDOST was overexpressed in patients with high grade, IDH wild type, 1p19q non-codel and MGMT un-methylated, which was associated with poor prognosis. Patients with high levels of DDOST, regardless of their clinical characteristics, had a worse prognosis. Immunohistochemical analysis confirmed the results of the above bioinformatics analysis. Mechanistic analysis revealed that DDOST was closely associated with the glioma microenvironment and negatively related to tumor-infiltrating B cells and CD4+ T cells and positively related to CAFs and tumor-associated macrophages. In conclusion, these findings suggested that DDOST mediated the immunosuppressive microenvironment of gliomas and could be an important biomarker in diagnosing and treating gliomas.
Keywords: DDOST; glioma; microenvironment; prognosis; progression.
Copyright © 2022 Chang, Pan, Zhao, Yan, Wang, Guo, Yang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

