Proteomic Signatures of Monocytes in Hereditary Recurrent Fevers
- PMID: 35812440
- PMCID: PMC9260596
- DOI: 10.3389/fimmu.2022.921253
Proteomic Signatures of Monocytes in Hereditary Recurrent Fevers
Abstract
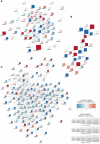
Hereditary periodic recurrent fevers (HRF) are monogenic autoinflammatory associated to mutations of some genes, such as diseases caused by mutations of including MEFV, TNFRSF1A and MVK genes. Despite the identification of the causative genes, the intracellular implications related to each gene variant are still largely unknown. A large -scale proteomic analysis on monocytes of these patients is aimed to identify with an unbiased approach the mean proteins and molecular interaction networks involved in the pathogenesis of these conditions. Monocytes from HRF 15 patients (5 with MFV, 5 TNFRSF1A and 5with MVK gene mutation) and 15 healthy donors (HDs) were analyzed by liquid chromatography and tandem mass spectrometry before and after lipopolysaccharide (LPS) stimulation. Significant proteins were analyzed through a Cytoscape analysis using the ClueGo app to identify molecular interaction networks. Protein networks for each HRF were performed through a STRING database analysis integrated with a DISEAE database query. About 5000 proteins for each HRF were identified. LPS treatment maximizes differences between up-regulated proteins in monocytes of HRF patients and HDs, independently from the disease's activity and ongoing treatments. Proteins significantly modulated in monocytes of the different HRF allowed creating a disease-specific proteomic signatures and interactive protein network. Proteomic analysis is able to dissect the different intracellular pathways involved in the inflammatory response of circulating monocytes in HRF patients. The present data may help to identify a "monocyte proteomic signature" for each condition and unravel new possible unexplored intracellular pathways possibly involved in their pathogenesis. These data will be also useful to identify possible differences and similarities between the different HRFs and some multifactorial recurrent fevers.
Keywords: Hereditary recurrent fevers; inflammation; monocytes; proteomic; signature.
Copyright © 2022 Penco, Petretto, Lavarello, Papa, Bertoni, Omenetti, Gueli, Finetti, Caorsi, Volpi and Gattorno.
Conflict of interest statement
FP reports speaker fee from SOBI. MG reports grants and personal fees from Novartis, grants and personal fees from SOBI, outside the submitted work. RP reports speaker fee from SOBI. AB reports speaker fee from SOBI. SV reports speaker fee from SOBI. RC reports speaker fee from SOBI. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

