Genetic Architecture of Powdery Mildew Resistance Revealed by a Genome-Wide Association Study of a Worldwide Collection of Flax (Linum usitatissimum L.)
- PMID: 35812909
- PMCID: PMC9263915
- DOI: 10.3389/fpls.2022.871633
Genetic Architecture of Powdery Mildew Resistance Revealed by a Genome-Wide Association Study of a Worldwide Collection of Flax (Linum usitatissimum L.)
Abstract
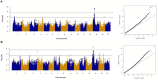
Powdery mildew is one of the most important diseases of flax and is particularly prejudicial to its yield and oil or fiber quality. This disease, caused by the obligate biotrophic ascomycete Oïdium lini, is progressing in France. Genetic resistance of varieties is critical for the control of this disease, but very few resistance genes have been identified so far. It is therefore necessary to identify new resistance genes to powdery mildew suitable to the local context of pathogenicity. For this purpose, we studied a worldwide diversity panel composed of 311 flax genotypes both phenotyped for resistance to powdery mildew resistance over 2 years of field trials in France and resequenced. Sequence reads were mapped on the CDC Bethune reference genome revealing 1,693,910 high-quality SNPs, further used for both population structure analysis and genome-wide association studies (GWASs). A number of four major genetic groups were identified, separating oil flax accessions from America or Europe and those from Asia or Middle-East and fiber flax accessions originating from Eastern Europe and those from Western Europe. A number of eight QTLs were detected at the false discovery rate threshold of 5%, located on chromosomes 1, 2, 4, 13, and 14. Taking advantage of the moderate linkage disequilibrium present in the flax panel, and using the available genome annotation, we identified potential candidate genes. Our study shows the existence of new resistance alleles against powdery mildew in our diversity panel, of high interest for flax breeding program.
Keywords: GBS; flax (Linum usitatissimum L.); genetic diversity; genome wide association studies (GWAS); powdery mildew.
Copyright © 2022 Speck, Trouvé, Enjalbert, Geffroy, Joets and Moreau.
Conflict of interest statement
AS and J-PT were employed by Terre De Lin. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Genome-wide association study and genomic selection of flax powdery mildew in Xinjiang Province.Front Plant Sci. 2024 May 28;15:1403276. doi: 10.3389/fpls.2024.1403276. eCollection 2024. Front Plant Sci. 2024. PMID: 38863531 Free PMC article.
-
History and prospects of flax genetic markers.Front Plant Sci. 2025 Jan 15;15:1495069. doi: 10.3389/fpls.2024.1495069. eCollection 2024. Front Plant Sci. 2025. PMID: 39881731 Free PMC article. Review.
-
Investigation of genetic diversity using molecular and biochemical markers associated with powdery mildew resistance in different flax (Linum usitatissimum L.) genotypes.BMC Plant Biol. 2024 May 17;24(1):412. doi: 10.1186/s12870-024-05113-5. BMC Plant Biol. 2024. PMID: 38760706 Free PMC article.
-
Insights into the Genetic Architecture and Genomic Prediction of Powdery Mildew Resistance in Flax (Linum usitatissimum L.).Int J Mol Sci. 2022 Apr 29;23(9):4960. doi: 10.3390/ijms23094960. Int J Mol Sci. 2022. PMID: 35563347 Free PMC article.
-
Molecular Advances to Combat Different Biotic and Abiotic Stresses in Linseed (Linum usitatissimum L.): A Comprehensive Review.Genes (Basel). 2023 Jul 17;14(7):1461. doi: 10.3390/genes14071461. Genes (Basel). 2023. PMID: 37510365 Free PMC article. Review.
Cited by
-
Genome-wide association study and genomic selection of flax powdery mildew in Xinjiang Province.Front Plant Sci. 2024 May 28;15:1403276. doi: 10.3389/fpls.2024.1403276. eCollection 2024. Front Plant Sci. 2024. PMID: 38863531 Free PMC article.
-
Overview and Management of the Most Common Eukaryotic Diseases of Flax (Linum usitatissimum).Plants (Basel). 2023 Jul 28;12(15):2811. doi: 10.3390/plants12152811. Plants (Basel). 2023. PMID: 37570965 Free PMC article. Review.
-
Key FAD2, FAD3, and SAD Genes Involved in the Fatty Acid Synthesis in Flax Identified Based on Genomic and Transcriptomic Data.Int J Mol Sci. 2023 Oct 4;24(19):14885. doi: 10.3390/ijms241914885. Int J Mol Sci. 2023. PMID: 37834335 Free PMC article.
-
History and prospects of flax genetic markers.Front Plant Sci. 2025 Jan 15;15:1495069. doi: 10.3389/fpls.2024.1495069. eCollection 2024. Front Plant Sci. 2025. PMID: 39881731 Free PMC article. Review.
-
Investigation of genetic diversity using molecular and biochemical markers associated with powdery mildew resistance in different flax (Linum usitatissimum L.) genotypes.BMC Plant Biol. 2024 May 17;24(1):412. doi: 10.1186/s12870-024-05113-5. BMC Plant Biol. 2024. PMID: 38760706 Free PMC article.
References
-
- Aly A. A., Mansour M. T. M., Mohamed H. I. (2017). Association of increase in some biochemical components with flax resistance to powdery mildew. Gesunde Pflanzen 69, 47–52. 10.1007/s10343-017-0387-7 - DOI
-
- Asgarinia P., Cloutier S., Duguid S., Rashid K., Mirlohi A., Banik M., et al. . (2013). Mapping quantitative trait loci for powdery mildew resistance in flax (Linum usitatissimum L.). Crop Sci. 53, 2462–2472. 10.2135/cropsci2013.05.0298 - DOI
LinkOut - more resources
Full Text Sources

