Keeping synapses in shape: degradation pathways in the healthy and aging brain
- PMID: 35813265
- PMCID: PMC9208270
- DOI: 10.1042/NS20210063
Keeping synapses in shape: degradation pathways in the healthy and aging brain
Abstract
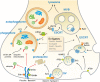
Synapses maintain their molecular composition, plasticity and function through the concerted action of protein synthesis and removal. The complex and polarized neuronal architecture poses specific challenges to the logistics of protein and organelle turnover since protein synthesis and degradation mainly happen in the cell soma. In addition, post-mitotic neurons accumulate damage over a lifetime, challenging neuronal degradative pathways and making them particularly susceptible to the effects of aging. This review will summarize the current knowledge on neuronal protein turnover mechanisms with a particular focus on the presynapse, including the proteasome, autophagy and the endolysosomal route and their roles in regulating presynaptic proteostasis and function. In addition, the author will discuss how physiological brain aging, which entails a progressive decline in cognitive functions, affects synapses and the degradative machinery.
Keywords: aging; autophagy; endolysosome; presynapse; proteostasis; ubiquitin proteasome system.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures
Similar articles
-
Autophagy and the endolysosomal system in presynaptic function.Cell Mol Life Sci. 2021 Mar;78(6):2621-2639. doi: 10.1007/s00018-020-03722-5. Epub 2020 Dec 19. Cell Mol Life Sci. 2021. PMID: 33340068 Free PMC article. Review.
-
Organization of Presynaptic Autophagy-Related Processes.Front Synaptic Neurosci. 2022 Mar 17;14:829354. doi: 10.3389/fnsyn.2022.829354. eCollection 2022. Front Synaptic Neurosci. 2022. PMID: 35368245 Free PMC article. Review.
-
Mechanism of synaptic protein turnover and its regulation by neuronal activity.Curr Opin Neurobiol. 2021 Aug;69:76-83. doi: 10.1016/j.conb.2021.02.006. Epub 2021 Mar 18. Curr Opin Neurobiol. 2021. PMID: 33744822 Review.
-
Homeostatic Roles of the Proteostasis Network in Dendrites.Front Cell Neurosci. 2020 Aug 14;14:264. doi: 10.3389/fncel.2020.00264. eCollection 2020. Front Cell Neurosci. 2020. PMID: 33013325 Free PMC article. Review.
-
The needs of a synapse-How local organelles serve synaptic proteostasis.EMBO J. 2022 Apr 4;41(7):e110057. doi: 10.15252/embj.2021110057. Epub 2022 Mar 14. EMBO J. 2022. PMID: 35285533 Free PMC article. Review.
Cited by
-
How do neurons age? A focused review on the aging of the microtubular cytoskeleton.Neural Regen Res. 2024 Sep 1;19(9):1899-1907. doi: 10.4103/1673-5374.390974. Epub 2023 Dec 15. Neural Regen Res. 2024. PMID: 38227514 Free PMC article.
-
Neuroplasticity to autophagy cross-talk in a therapeutic effect of physical exercises and irisin in ADHD.Front Mol Neurosci. 2023 Jan 26;15:997054. doi: 10.3389/fnmol.2022.997054. eCollection 2022. Front Mol Neurosci. 2023. PMID: 36776770 Free PMC article. Review.
-
Abl depletion via autophagy mediates the beneficial effects of quercetin against Alzheimer pathology across species.Cell Death Discov. 2023 Oct 14;9(1):376. doi: 10.1038/s41420-023-01592-x. Cell Death Discov. 2023. PMID: 37838776 Free PMC article.
-
Celebrating 125 years of the synapse: from Sherrington to the present day.Neuronal Signal. 2022 Dec 22;6(4):NS20220015. doi: 10.1042/NS20220015. eCollection 2022 Dec. Neuronal Signal. 2022. PMID: 36618960 Free PMC article.
-
"Dirty Dancing" of Calcium and Autophagy in Alzheimer's Disease.Life (Basel). 2023 May 15;13(5):1187. doi: 10.3390/life13051187. Life (Basel). 2023. PMID: 37240832 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

