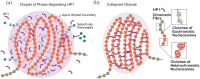
HP1-Driven Micro-Phase Separation of Heterochromatin-Like Domains/Complexes
- PMID: 35813402
- PMCID: PMC9260563
- DOI: 10.1177/25168657221109766
HP1-Driven Micro-Phase Separation of Heterochromatin-Like Domains/Complexes
Keywords: H3K9me3; HP1; Heterochromatin; Hi-C; phase separation.
Conflict of interest statement
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Figures

References
-
- Machida S, Takizawa Y, Ishimaru M, et al.. Structural basis of heterochromatin formation by human HP1. Mol Cell. 2018;69:385-397.e8. - PubMed
LinkOut - more resources
Full Text Sources

