Novel Insights Into the Phylogeny and Biotechnological Potential of Weissella Species
- PMID: 35814678
- PMCID: PMC9257631
- DOI: 10.3389/fmicb.2022.914036
Novel Insights Into the Phylogeny and Biotechnological Potential of Weissella Species
Abstract
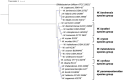
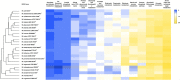
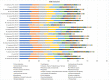
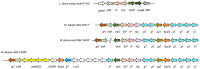
In this study, the genomes of the Weissella (W.) beninensis, W. diestrammenae, W. fabalis, W. fabaria, W. ghanensis, and W. uvarum type strains were sequenced and analyzed. Moreover, the ability of these strains to metabolize 95 carbohydrates was investigated, and the genetic determinants of such capability were searched within the sequenced genomes. 16S rRNA gene and genome-based-phylogeny of all the Weissella species described to date allowed a reassessment of the Weissella genus species groups. As a result, six distinct species groups within the genus, namely, W. beninensis, W. kandleri, W. confusa, W. halotolerans, W. oryzae, and W. paramesenteroides species groups, could be described. Phenotypic analyses provided further knowledge about the ability of the W. beninensis, W. ghanensis, W. fabaria, W. fabalis, W. uvarum, and W. diestrammenae type strains to metabolize certain carbohydrates and confirmed the interspecific diversity of the analyzed strains. Moreover, in many cases, the carbohydrate metabolism pathway and phylogenomic species group clustering overlapped. The novel insights provided in our study significantly improved the knowledge about the Weissella genus and allowed us to identify features that define the role of the analyzed type strains in fermentative processes and their biotechnological potential.
Keywords: Weissella spp.; carbohydrate metabolism; genomics; phylogenomics; trehalose.
Copyright © 2022 Fanelli, Montemurro, Chieffi, Cho, Franz, Dell'Aquila, Rizzello and Fusco.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Probiotic Potential and Safety Assessment of Type Strains of Weissella and Periweissella Species.Microbiol Spectr. 2023 Feb 27;11(2):e0304722. doi: 10.1128/spectrum.03047-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36847557 Free PMC article.
-
Motility in Periweissella Species: Genomic and Phenotypic Characterization and Update on Motility in Lactobacillaceae.Microorganisms. 2023 Dec 5;11(12):2923. doi: 10.3390/microorganisms11122923. Microorganisms. 2023. PMID: 38138067 Free PMC article.
-
The Weissella Genus: Clinically Treatable Bacteria with Antimicrobial/Probiotic Effects on Inflammation and Cancer.Microorganisms. 2022 Dec 7;10(12):2427. doi: 10.3390/microorganisms10122427. Microorganisms. 2022. PMID: 36557680 Free PMC article. Review.
-
Weissella cryptocerci sp. nov., isolated from gut of the insect Cryptocercus kyebangensis.Int J Syst Evol Microbiol. 2019 Sep;69(9):2801-2806. doi: 10.1099/ijsem.0.003564. Int J Syst Evol Microbiol. 2019. PMID: 31246166
-
The genus Weissella: taxonomy, ecology and biotechnological potential.Front Microbiol. 2015 Mar 17;6:155. doi: 10.3389/fmicb.2015.00155. eCollection 2015. Front Microbiol. 2015. PMID: 25852652 Free PMC article. Review.
Cited by
-
Probiotic Potential and Safety Assessment of Type Strains of Weissella and Periweissella Species.Microbiol Spectr. 2023 Feb 27;11(2):e0304722. doi: 10.1128/spectrum.03047-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36847557 Free PMC article.
-
Antimicrobial Activity of Probiotic Bacteria Isolated from Plants: A Review.Foods. 2025 Feb 4;14(3):495. doi: 10.3390/foods14030495. Foods. 2025. PMID: 39942088 Free PMC article. Review.
-
Motility in Periweissella Species: Genomic and Phenotypic Characterization and Update on Motility in Lactobacillaceae.Microorganisms. 2023 Dec 5;11(12):2923. doi: 10.3390/microorganisms11122923. Microorganisms. 2023. PMID: 38138067 Free PMC article.
-
Swine farm environmental microbiome: exploring microbial ecology and functionality across farms with high and low sanitary status.Front Vet Sci. 2024 Jul 3;11:1401561. doi: 10.3389/fvets.2024.1401561. eCollection 2024. Front Vet Sci. 2024. PMID: 39021414 Free PMC article.
-
The Weissella and Periweissella genera: up-to-date taxonomy, ecology, safety, biotechnological, and probiotic potential.Front Microbiol. 2023 Dec 11;14:1289937. doi: 10.3389/fmicb.2023.1289937. eCollection 2023. Front Microbiol. 2023. PMID: 38169702 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

