Overexpression of the Mitochondrial Malic Enzyme Genes (malC and malD) Improved the Lipid Accumulation in Mucor circinelloides WJ11
- PMID: 35814694
- PMCID: PMC9260706
- DOI: 10.3389/fmicb.2022.919364
Overexpression of the Mitochondrial Malic Enzyme Genes (malC and malD) Improved the Lipid Accumulation in Mucor circinelloides WJ11
Abstract
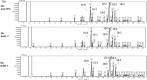
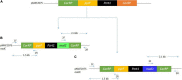
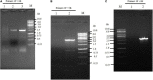
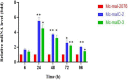
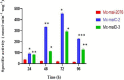
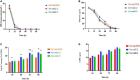
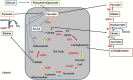
Mucor circinelloides serves as a model organism to investigate the lipid metabolism in oleaginous microorganisms. It is considered as an important producer of γ-linolenic acid (GLA) that has vital medicinal benefits. In this study, we used WJ11, a high lipid-producing strain of M. circinelloides (36% w/w lipid, cell dry weight, CDW), to examine the role in lipid accumulation of two mitochondrial malic enzyme (ME) genes malC and malD. The homologous overexpression of both malC and malD genes enhanced the total lipid content of WJ11 by 41.16 and 32.34%, respectively. In parallel, the total content of GLA was enhanced by 16.73 and 46.76% in malC and malD overexpressing strains, respectively, because of the elevation of total lipid content. The fact that GLA content was enhanced more in the strain with lower lipid content increase and vice versa, indicated that engineering of mitochondrial MEs altered the fatty acid profile. Our results reveal that mitochondrial ME plays an important role in lipid metabolism and suggest that future approaches may involve simultaneous overexpression of distinct ME genes to boost lipid accumulation even further.
Keywords: Mucor circinelloides; genetic engineering; health benefits; lipid accumulation; malic enzyme; oleaginous fungi; γ-linolenic acid.
Copyright © 2022 Fazili, Shah, Albeshr, Naz, Dar, Yang, Garre, Fazili, Bhat and Song.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Role of Cytosolic Malic Enzyme in Oleaginicity of High-Lipid-Producing Fungal Strain Mucor circinelloides WJ11.J Fungi (Basel). 2022 Mar 5;8(3):265. doi: 10.3390/jof8030265. J Fungi (Basel). 2022. PMID: 35330267 Free PMC article.
-
Role of Snf-β in lipid accumulation in the high lipid-producing fungus Mucor circinelloides WJ11.Microb Cell Fact. 2021 Feb 27;20(1):52. doi: 10.1186/s12934-021-01545-y. Microb Cell Fact. 2021. PMID: 33639948 Free PMC article.
-
Simultaneous overexpression of ∆6-, ∆12- and ∆9-desaturases enhanced the production of γ-linolenic acid in Mucor circinelloides WJ11.Front Microbiol. 2022 Dec 15;13:1078157. doi: 10.3389/fmicb.2022.1078157. eCollection 2022. Front Microbiol. 2022. PMID: 36590442 Free PMC article.
-
Mucor circinelloides: a model organism for oleaginous fungi and its potential applications in bioactive lipid production.Microb Cell Fact. 2022 Feb 28;21(1):29. doi: 10.1186/s12934-022-01758-9. Microb Cell Fact. 2022. PMID: 35227264 Free PMC article. Review.
-
Regulation of lipid accumulation in oleaginous micro-organisms.Biochem Soc Trans. 2002 Nov;30(Pt 6):1047-50. doi: 10.1042/bst0301047. Biochem Soc Trans. 2002. PMID: 12440969 Review.
Cited by
-
Oleaginous fungi: a promising source of biofuels and nutraceuticals with enhanced lipid production strategies.Arch Microbiol. 2024 Jul 2;206(7):338. doi: 10.1007/s00203-024-04054-9. Arch Microbiol. 2024. PMID: 38955856 Review.
-
Metabolic engineering of Mucor circinelloides to improve astaxanthin production.World J Microbiol Biotechnol. 2024 Nov 2;40(12):374. doi: 10.1007/s11274-024-04181-x. World J Microbiol Biotechnol. 2024. PMID: 39487367
-
Malic enzymes in cancer: Regulatory mechanisms, functions, and therapeutic implications.Redox Biol. 2024 Sep;75:103273. doi: 10.1016/j.redox.2024.103273. Epub 2024 Jul 19. Redox Biol. 2024. PMID: 39142180 Free PMC article. Review.
-
AoMae1 Regulates Hyphal Fusion, Lipid Droplet Accumulation, Conidiation, and Trap Formation in Arthrobotrys oligospora.J Fungi (Basel). 2023 Apr 21;9(4):496. doi: 10.3390/jof9040496. J Fungi (Basel). 2023. PMID: 37108952 Free PMC article.
References
-
- Chaney A. L., Marbach E. P. (1962). Modified reagents for determination of urea and ammonia. Clin. Chem. 8 130–132. - PubMed
-
- Dulermo T., Lazar Z., Dulermo R., Rakicka M., Haddouche R., Nicaud J.-M. (2015). Analysis of ATP-citrate lyase and malic enzyme mutants of Yarrowia lipolytica points out the importance of mannitol metabolism in fatty acid synthesis. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 1851 1107–1117. 10.1016/j.bbalip.2015.04.007 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

