CLEC10A can serve as a potential therapeutic target and its level correlates with immune infiltration in breast cancer
- PMID: 35814828
- PMCID: PMC9260715
- DOI: 10.3892/ol.2022.13405
CLEC10A can serve as a potential therapeutic target and its level correlates with immune infiltration in breast cancer
Abstract
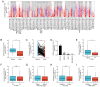
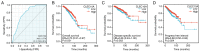
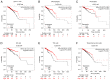
Breast cancer (BC) is one of the most common malignant cancers in females worldwide and greatly threatens women's health. The C-type lectin domain family 10 member A (CLEC10A) is a member of the C-type lectin receptor family that has been previously reported to promote the antitumor activity of immune cells. In the present study, the potential prognostic value of CLEC10A expression in BC was assessed using data from The Cancer Genome Atlas online database. Differences in the mRNA expression levels of CLEC10A between BC and normal tissues were then analyzed using the Tumor Immune Estimation Resource (TIMER) platform and the University of Alabama at Birmingham Cancer data analysis portal. Reverse transcription-quantitative PCR was performed to validate the results of this analysis. The Kaplan-Meier plotter database was used to evaluate the association between the mRNA expression levels of CLEC10A and clinical prognosis of BC. Based on the association between the mRNA expression levels of CLEC10A and the tumor immune microenvironment, the TIMER platform and the Tumor and Immune System Interaction Database website were utilized to assess the correlation between CLEC10A expression and the degree of tumor immune cell infiltration. The present study revealed that CLEC10A expression was significantly lower in BC tissues compared with that in normal tissues, which was in turn associated with poorer clinical outcomes. This suggested that lower CLEC10A expression levels were associated with unfavorable prognosis in BC. In addition, the expression level of CLEC10A was found to be positively associated with the level of different tumor-infiltrating immune cells in BC, including CD8 T cells, B cells, macrophages and NK cells which, was in turn closely correlated with some gene markers such as CD19, CD8A, KIR2DS4 and PTGS2. These results suggest that the relationship between lower CLEC10A expression level and poor prognosis in BC may be due to the role of CLEC10A in the tumor immune microenvironment. In conclusion, CLEC10A may be a potential biomarker that can be used to efficiently predict prognosis in patients with BC.
Keywords: C-type lectin domain family 10 member A; biomarkers; breast cancer; immune cell; overall survival; prognosis.
Copyright: © Tang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Immunological role and prognostic potential of CLEC10A in pan-cancer.Am J Transl Res. 2022 May 15;14(5):2844-2860. eCollection 2022. Am J Transl Res. 2022. PMID: 35702069 Free PMC article.
-
The prognostic significance of KLRB1 and its further association with immune cells in breast cancer.PeerJ. 2023 Jul 24;11:e15654. doi: 10.7717/peerj.15654. eCollection 2023. PeerJ. 2023. PMID: 37520246 Free PMC article.
-
CLEC10A is a prognostic biomarker and correlated with clinical pathologic features and immune infiltrates in lung adenocarcinoma.J Cell Mol Med. 2021 Apr;25(7):3391-3399. doi: 10.1111/jcmm.16416. Epub 2021 Mar 2. J Cell Mol Med. 2021. PMID: 33655701 Free PMC article.
-
High expression of CLEC10A in head and neck squamous cell carcinoma indicates favorable prognosis and high-level immune infiltration status.Cell Immunol. 2022 Feb;372:104472. doi: 10.1016/j.cellimm.2021.104472. Epub 2021 Dec 22. Cell Immunol. 2022. PMID: 35093731
-
The pleiotropic CLEC10A: implications for harnessing this receptor in the tumor microenvironment.Expert Opin Ther Targets. 2024 Jul;28(7):601-612. doi: 10.1080/14728222.2024.2374743. Epub 2024 Jul 3. Expert Opin Ther Targets. 2024. PMID: 38946482 Review.
Cited by
-
Evaluation of the efficacy of totally implantable venous access port in breast cancer patients undergoing chemotherapy.Discov Oncol. 2025 Mar 24;16(1):383. doi: 10.1007/s12672-025-02020-5. Discov Oncol. 2025. PMID: 40126722 Free PMC article.
-
Elevated BEAN1 expression correlates with poor prognosis, immune evasion, and chemotherapy resistance in rectal adenocarcinoma.Discov Oncol. 2024 Sep 14;15(1):446. doi: 10.1007/s12672-024-01321-5. Discov Oncol. 2024. PMID: 39276259 Free PMC article.
-
SADLN: Self-attention based deep learning network of integrating multi-omics data for cancer subtype recognition.Front Genet. 2023 Jan 4;13:1032768. doi: 10.3389/fgene.2022.1032768. eCollection 2022. Front Genet. 2023. PMID: 36685873 Free PMC article.
-
Mannose and Lactobionic Acid in Nasal Vaccination: Enhancing Antigen Delivery via C-Type Lectin Receptors.Pharmaceutics. 2024 Oct 8;16(10):1308. doi: 10.3390/pharmaceutics16101308. Pharmaceutics. 2024. PMID: 39458637 Free PMC article. Review.
-
Unveiling the hidden role of SDHA in breast cancer proliferation: a novel therapeutic avenue.Cancer Cell Int. 2025 Mar 21;25(1):108. doi: 10.1186/s12935-025-03746-6. Cancer Cell Int. 2025. PMID: 40119440 Free PMC article.
References
-
- Saad ED, Squifflet P, Burzykowski T, Quinaux E, Delaloge S, Mavroudis D, Perez E, Piccart-Gebhart M, Schneider BP, Slamon D, et al. Disease-free survival as a surrogate for overall survival in patients with HER2-positive, early breast cancer in trials of adjuvant trastuzumab for up to 1 year: A systematic review and meta-analysis. Lancet Oncol. 2019;20:361–370. doi: 10.1016/S1470-2045(18)30750-2. - DOI - PMC - PubMed
-
- Shachar SS, Deal AM, Weinberg M, Williams GR, Nyrop KA, Popuri K, Choi SK, Muss HB. Body composition as a predictor of toxicity in patients receiving anthracycline and taxane-based chemotherapy for early-stage breast cancer. Clin Cancer Res. 2017;23:3537–3543. doi: 10.1158/1078-0432.CCR-16-0940. - DOI - PMC - PubMed
-
- Lischka A, Doberstein N, Freitag-Wolf S, Kocak A, Gemoll T, Heselmeyer-Haddad K, Ried T, Auer G, Habermann JK. Genome instability profiles predict disease outcome in a cohort of 4,003 patients with breast cancer. Clin Cancer Res. 2020;26:4606–4615. doi: 10.1158/1078-0432.CCR-20-0566. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
