The Complete Chloroplast Genome Sequence of Cicer bijugum, Genome Organization, and Comparison with Related Species
- PMID: 35814936
- PMCID: PMC9199535
- DOI: 10.2174/1389202923666220211113708
The Complete Chloroplast Genome Sequence of Cicer bijugum, Genome Organization, and Comparison with Related Species
Abstract
Background: Chickpea is one of Turkey's most significant legumes, and because of its high nutritional value, it is frequently preferred in human nourishment.Chloroplasts, which have their own genetic material, are organelles responsible for photosynthesis in plant cells and their genome contains non-trivial information about the molecular features and evolutionary process of plants.
Objective: Current study aimed at revealing complete chloroplast genome sequence of one of the wild type Cicer species, Cicer bijugum, and comparing its genome with cultivated Cicer species, Cicer arietinum, by using bioinformatics analysis tools. Except for Cicer arietinum, there has been no study on the chloroplast genome sequence of Cicer species.Therefore, we targeted to reveal the complete chloroplast genome sequence of wild type Cicer species, Cicer bijugum, and compare the chloroplast genome of Cicer bijugum with the cultivated one Cicer arietinum.
Methods: In this study, we sequenced the whole chloroplast genome of Cicer bijugum, one of the wild types of chickpea species, with the help Next Generation Sequencing platform and compared it with the chloroplast genome of the cultivated chickpea species, Cicer arietinum, by using online bioinformatics analysis tools.
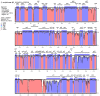
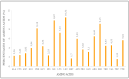
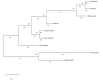
Results: We determined the size of the chloroplast genome of C. bijugum as 124,804 bp and found that C. bijugum did not contain an inverted repeat region in its chloroplast genome. Comparative analysis of the C. bijugum chloroplast genome uncovered thirteen hotspot regions (psbA, matK, rpoB, rpoC1, rpoC2, psbI, psbK, accD, rps19, ycf2, ycf1, rps15, and ndhF) and seven of them (matK, accD, rps19, ycf1, ycf2, rps15 and ndhF) could potentially be used as strong molecular markers for species identification. It has been determined that C. bijugum was phylogenetically closer to cultivated chickpea as compared to the other species.
Conclusion: It is aimed that the data obtained from this study, which is the first study in which whole chloroplast genomes of wild chickpea species were sequenced, will guide researchers in future molecular, evolutionary, and genetic engineering studies with chickpea species.
Keywords: Cicer bijugum; Wild type chickpea; bioinformatics; chloroplast genome; comparative genome analysis; genome organization.
© 2022 Bentham Science Publishers.
Figures





Similar articles
-
The complete chloroplast genome of Cicer reticulatum and comparative analysis against relative Cicer species.Sci Rep. 2023 Oct 19;13(1):17871. doi: 10.1038/s41598-023-44599-1. Sci Rep. 2023. PMID: 37857674 Free PMC article.
-
Characterization of Seed Proteome Profile of Wild and Cultivated Chickpeas of India.Protein Pept Lett. 2021;28(3):323-332. doi: 10.2174/0929866527666200910164118. Protein Pept Lett. 2021. PMID: 32914710
-
Proteolytic Activity in the Midgut of Helicoverpa armigera (Noctuidae: Lepidoptera) Larvae Fed on Wild Relatives of Chickpea, Cicer arietinum.J Econ Entomol. 2018 Sep 26;111(5):2409-2415. doi: 10.1093/jee/toy160. J Econ Entomol. 2018. PMID: 29924350
-
Chickpea improvement: role of wild species and genetic markers.Biotechnol Genet Eng Rev. 2008;25:267-313. doi: 10.5661/bger-25-267. Biotechnol Genet Eng Rev. 2008. PMID: 21412359 Review.
-
Nutritional composition, health benefits and bio-active compounds of chickpea (Cicer arietinum L.).Front Nutr. 2023 Sep 28;10:1218468. doi: 10.3389/fnut.2023.1218468. eCollection 2023. Front Nutr. 2023. PMID: 37854353 Free PMC article. Review.
Cited by
-
The complete chloroplast genome of Cicer reticulatum and comparative analysis against relative Cicer species.Sci Rep. 2023 Oct 19;13(1):17871. doi: 10.1038/s41598-023-44599-1. Sci Rep. 2023. PMID: 37857674 Free PMC article.
-
Insights into Phylogeny, Taxonomy, Origins and Evolution of Crataegus and Mespilus, Based on Comparative Chloroplast Genome Analysis.Genes (Basel). 2025 Feb 7;16(2):204. doi: 10.3390/genes16020204. Genes (Basel). 2025. PMID: 40004533 Free PMC article.
-
Comparative Plastome Analysis Between Endangered Mangrove Species Acanthus ebracteatus and Acanthus Relatives Provides Insights into Its Origin and Adaptive Evolution.Ecol Evol. 2024 Nov 20;14(11):e70566. doi: 10.1002/ece3.70566. eCollection 2024 Nov. Ecol Evol. 2024. PMID: 39568763 Free PMC article.
-
Comparison of organelle genomes between endangered mangrove plant Dolichandrone spathacea to terrestrial relative provides insights into its origin and adaptative evolution.Front Plant Sci. 2024 Sep 23;15:1442178. doi: 10.3389/fpls.2024.1442178. eCollection 2024. Front Plant Sci. 2024. PMID: 39376234 Free PMC article.
-
Comparative Analysis of Plastome Sequences of Seven Tulipa L. (Liliaceae Juss.) Species from Section Kolpakowskianae Raamsd. Ex Zonn and Veldk.Int J Mol Sci. 2024 Jul 18;25(14):7874. doi: 10.3390/ijms25147874. Int J Mol Sci. 2024. PMID: 39063115 Free PMC article.
References
-
- Lei X., Zhou Q., Li W., Qin G., Shen X., Zhang N. Stilbenoids from leguminosae and their bioactivities. Med. Res. 2019;3:200004. doi: 10.21127/yaoyimr20200004. - DOI
-
- Abbasi B.A., Iqbal J., Mahmood T. Assessment of phylogenetics relationship among the selected species of family leguminosae based on chloroplast rps14 gene. Pak. J. Bot. 2021;53:1307–1313. doi: 10.30848/PJB2021-4(22). - DOI
-
- Obistioiu D., Cocan I., Tîrziu E., Herman V., Negrea M., Cucerzan A., Neacsu A.G., Cozma A.L., Nichita I., Hulea A., Radulov I., Alexa E. Phytochemical profile and microbiological activity of some plants belonging to the fabaceae family. Antibiotics (Basel) 2021;10(6):662. doi: 10.3390/antibiotics10060662. - DOI - PMC - PubMed
-
- Oyebanji O.O., Salako G., Nneji L.M., Oladipo S.O., Bolarinwa K.A., Chukwuma E.C., et al. Impact of climate change on the spatial distribution of endemic legume species of the Guineo-Congolian forest, Africa. Ecol. Indic. 2021;122:107282. doi: 10.1016/j.ecolind.2020.107282. - DOI
-
- Alloosh M., Hamwieh A., Ahmed S., Alkai B. Genetic diversity of Fusarium oxysporum f. sp. Ciceris isolates affecting chickpea in Syria. Crop Prot. 2019;124 doi: 10.1016/j.cropro.2019.104863. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous
