Novel MicroRNAs and their Functional Targets from Phytophthora infestans and Phytophthora cinnamomi
- PMID: 35814938
- PMCID: PMC9199537
- DOI: 10.2174/1389202923666211223122305
Novel MicroRNAs and their Functional Targets from Phytophthora infestans and Phytophthora cinnamomi
Abstract
Background: Even though miRNAs play vital roles in developmental biology by regulating the translation of mRNAs, they are poorly studied in oomycetes, especially in the plant pathogen Phytophthora.
Objective: The study aimed to predict and identify the putative miRNAs and their targets in Phytophthora infestans and Phytophthora cinnamomi.
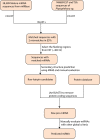
Methods: The homology-based comparative method was used to identify the unique miRNA sequences in P. infestans and P. cinnamomi with 148,689 EST and TSA sequences of these species. Secondary structure prediction of sRNAs for the 76 resultant sequences has been performed with the MFOLD tool, and their targets were predicted using psRNATarget.
Results: Novel miRNAs, miR-8210 and miR-4968, were predicted from P. infestans and P. cinnamomi, respectively, along with their structural features. The newly identified miRNAs were identified to play important roles in gene regulation, with few of their target genes predicted as transcription factors, tumor suppressor genes, stress-responsive genes, DNA repair genes, etc.
Conclusion: The miRNAs and their targets identified have opened new interference and editing targets for the development of Phytophthora resistant crop varieties.
Keywords: Data mining; fungus; genome annotation; miRNA; oomycete; siRNA; target prediction.
© 2022 Bentham Science Publishers.
Figures
Similar articles
-
Identification of miRNAs Involving Potato-Phytophthora infestans Interaction.Plants (Basel). 2023 Jan 19;12(3):461. doi: 10.3390/plants12030461. Plants (Basel). 2023. PMID: 36771544 Free PMC article.
-
Computational and comparative analyses of 150 full-length cDNA sequences from the oomycete plant pathogen Phytophthora infestans.Fungal Genet Biol. 2006 Jan;43(1):20-33. doi: 10.1016/j.fgb.2005.10.003. Epub 2005 Dec 27. Fungal Genet Biol. 2006. PMID: 16380277
-
Draft genomes of two Australian strains of the plant pathogen, Phytophthora cinnamomi.F1000Res. 2017 Nov 8;6:1972. doi: 10.12688/f1000research.12867.2. eCollection 2017. F1000Res. 2017. PMID: 29188023 Free PMC article.
-
Phytopathogenic oomycetes: a review focusing on Phytophthora cinnamomi and biotechnological approaches.Mol Biol Rep. 2020 Nov;47(11):9179-9188. doi: 10.1007/s11033-020-05911-8. Epub 2020 Oct 17. Mol Biol Rep. 2020. PMID: 33068230 Review.
-
In silico characterization of molecular factors involved in metabolism and pathogenicity of Phytophthora cinnamomi.Mol Biol Rep. 2022 Feb;49(2):1463-1473. doi: 10.1007/s11033-021-06901-0. Epub 2021 Nov 9. Mol Biol Rep. 2022. PMID: 34751913 Review.
References
-
- Moore N., Barrett S., Bowen B., Shearer B., Hardy G. The role of fire on Phytophthora dieback caused by the root pathogen Phytophthora cinnamomi in the Stirling Range National Park, Western Australia.; In: Proc. 11th International Mediterranean Ecosystems (MEDECOS) Conference; 2007 Sep 2-5; Perth, Western Australia. pp. 165–166.
-
- Scott P., Bader M.K., Burgess T., Hardy G., Williams N. Global biogeography and invasion risk of the plant pathogen genus Phytophthora. Environ. Sci. Policy. 2019;101:175–182. doi: 10.1016/j.envsci.2019.08.020. - DOI
LinkOut - more resources
Full Text Sources
Research Materials