A Combinational Approach for More Efficient miRNA Biosensing
- PMID: 35814939
- PMCID: PMC9199536
- DOI: 10.2174/1389202923666220204160912
A Combinational Approach for More Efficient miRNA Biosensing
Abstract
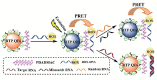
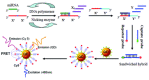
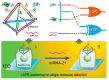
MicroRNAs, short single-stranded noncoding RNAs ranging in length from 18 ~ 24 bp, are found in all kingdoms of eukaryotes and even viruses. It was found that miRNAs are involved in a variety of biological processes, and their intracellular aberrant expression is related to diseases and abnormalities in the immune system. Since then, it has been considered essential to develop an efficient miRNA detection system. In this review, the limitations of traditional scheme-based miRNA detection methods are compared and analyzed. In particular, nucleic acid amplification-based miRNA detection methods and nanomaterial-based miRNA detection methods, which are widely used as a biosensing platform because of various features and advantages, such as high sensitivity, specificity, and simplicity, are analyzed. Based on this analysis, the latest examples of a combination of the advantages of nucleic acid amplification and those of nanomaterials are examined to suggest the characteristics of the next-generation miRNA biosensing.
Keywords: biosensing platform; miRNA detection system; miRNAs; nanomaterial-based miRNA; next-generation miRNA biosensing; nucleic acid amplification.
© 2022 Bentham Science Publishers.
Figures

Similar articles
-
Research advances in the detection of miRNA.J Pharm Anal. 2019 Aug;9(4):217-226. doi: 10.1016/j.jpha.2019.05.004. Epub 2019 May 25. J Pharm Anal. 2019. PMID: 31452959 Free PMC article. Review.
-
Recent Progress in Nanomaterials Modified Electrochemical Biosensors for the Detection of MicroRNA.Micromachines (Basel). 2021 Nov 17;12(11):1409. doi: 10.3390/mi12111409. Micromachines (Basel). 2021. PMID: 34832823 Free PMC article. Review.
-
Isothermal Amplification for MicroRNA Detection: From the Test Tube to the Cell.Acc Chem Res. 2017 Apr 18;50(4):1059-1068. doi: 10.1021/acs.accounts.7b00040. Epub 2017 Mar 29. Acc Chem Res. 2017. PMID: 28355077 Review.
-
PCDetection: PolyA-CRISPR/Cas12a-based miRNA detection without PAM restriction.Biosens Bioelectron. 2022 Oct 15;214:114497. doi: 10.1016/j.bios.2022.114497. Epub 2022 Jun 30. Biosens Bioelectron. 2022. PMID: 35797934
-
Target-swiped DNA lock for electrochemical sensing of miRNAs based on DNAzyme-assisted primer-generation amplification.Mikrochim Acta. 2021 Jul 15;188(8):255. doi: 10.1007/s00604-021-04815-z. Mikrochim Acta. 2021. PMID: 34264390
Cited by
-
Innovations in microRNA-based electrochemical biosensors for essential hypertension.Hypertens Res. 2024 Jul;47(7):2003-2005. doi: 10.1038/s41440-024-01728-1. Epub 2024 May 17. Hypertens Res. 2024. PMID: 38760525 No abstract available.
References
Publication types
LinkOut - more resources
Full Text Sources
